
Human papillomavirus 199
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; Gammapapillomavirus 12
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
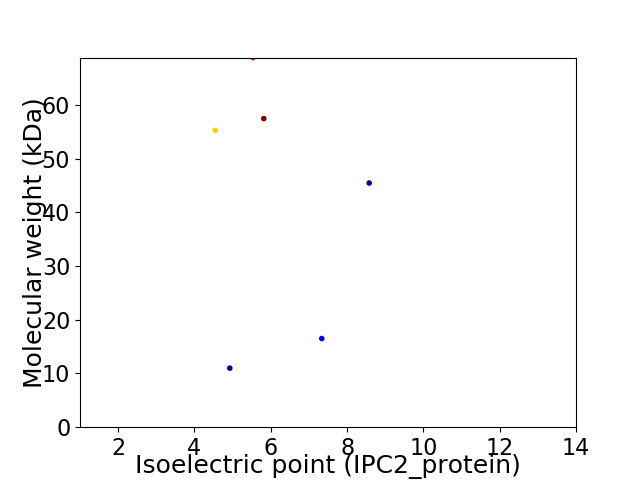
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A0N9E2S0|A0A0N9E2S0_9PAPI Minor capsid protein L2 OS=Human papillomavirus 199 OX=1545700 GN=L2 PE=3 SV=1
MM1 pKa = 7.54SATTRR6 pKa = 11.84RR7 pKa = 11.84KK8 pKa = 9.3RR9 pKa = 11.84AAPVDD14 pKa = 4.38LYY16 pKa = 11.1KK17 pKa = 10.86SCLAGGDD24 pKa = 4.48CITDD28 pKa = 3.25VKK30 pKa = 11.4NKK32 pKa = 10.28FEE34 pKa = 4.24NTTLADD40 pKa = 3.09ILLKK44 pKa = 10.81VFGSIVYY51 pKa = 10.18FGGLGIGSGKK61 pKa = 10.63GSGGSLGYY69 pKa = 10.1RR70 pKa = 11.84PLGSEE75 pKa = 3.82PTTGRR80 pKa = 11.84VAPTAPVRR88 pKa = 11.84PSVIVDD94 pKa = 3.32PLVPPDD100 pKa = 4.09VITVEE105 pKa = 4.34PSAPSIIPLAEE116 pKa = 3.89GRR118 pKa = 11.84PNIDD122 pKa = 2.96FAAPDD127 pKa = 3.92GGPGLGAEE135 pKa = 4.46EE136 pKa = 4.36IEE138 pKa = 5.03LYY140 pKa = 9.76TIKK143 pKa = 10.12TPTTDD148 pKa = 2.55IGGVGSEE155 pKa = 4.0PTVISSEE162 pKa = 4.14EE163 pKa = 3.99GATAILPVDD172 pKa = 5.13PIPEE176 pKa = 3.87RR177 pKa = 11.84PAQVFYY183 pKa = 11.21DD184 pKa = 4.41PSAPSQYY191 pKa = 10.45EE192 pKa = 3.87LNIFAAHH199 pKa = 7.36PSTSSDD205 pKa = 2.67INVFVDD211 pKa = 3.33NSFSGKK217 pKa = 9.66VVGTFEE223 pKa = 4.86EE224 pKa = 4.74IPLEE228 pKa = 4.2KK229 pKa = 10.67YY230 pKa = 10.39NYY232 pKa = 8.11STFEE236 pKa = 3.81IEE238 pKa = 4.53EE239 pKa = 4.6PPSTSTPTRR248 pKa = 11.84SLEE251 pKa = 4.13RR252 pKa = 11.84VATRR256 pKa = 11.84AKK258 pKa = 10.46ALYY261 pKa = 10.44NKK263 pKa = 7.67FTRR266 pKa = 11.84QVSVKK271 pKa = 10.46DD272 pKa = 3.88PNFLLQPSRR281 pKa = 11.84LVQFEE286 pKa = 4.12FDD288 pKa = 3.54NPAFDD293 pKa = 5.43RR294 pKa = 11.84DD295 pKa = 3.46VTLQFEE301 pKa = 4.27QDD303 pKa = 3.75VQQVTAAPNPDD314 pKa = 3.59FADD317 pKa = 4.37VIQLGRR323 pKa = 11.84PILSQTEE330 pKa = 4.04EE331 pKa = 4.17GVVRR335 pKa = 11.84VSRR338 pKa = 11.84LGKK341 pKa = 9.83VGTVATRR348 pKa = 11.84SGRR351 pKa = 11.84VIGQPVHH358 pKa = 6.04YY359 pKa = 9.4FQDD362 pKa = 3.51LSVIEE367 pKa = 4.36PEE369 pKa = 4.28SIEE372 pKa = 3.74LHH374 pKa = 6.18GFAEE378 pKa = 4.75HH379 pKa = 6.97SNQTTIVDD387 pKa = 4.24DD388 pKa = 4.53LLSSSTFQNPVFDD401 pKa = 4.46SNAVFNEE408 pKa = 3.5DD409 pKa = 4.76LLLDD413 pKa = 4.65EE414 pKa = 4.87YY415 pKa = 11.86NEE417 pKa = 4.8DD418 pKa = 3.57FGNTNLVVATTDD430 pKa = 3.22EE431 pKa = 4.4AEE433 pKa = 4.26EE434 pKa = 4.33TFSFPILAPSPSVKK448 pKa = 10.3VFVTDD453 pKa = 3.1VGNIFVTSSKK463 pKa = 10.16DD464 pKa = 3.14AHH466 pKa = 5.57STITVLNTDD475 pKa = 5.08LVPLTPSIITSSYY488 pKa = 9.36NNFYY492 pKa = 10.92LEE494 pKa = 3.96PFYY497 pKa = 11.28VPRR500 pKa = 11.84KK501 pKa = 8.28KK502 pKa = 10.14RR503 pKa = 11.84RR504 pKa = 11.84RR505 pKa = 11.84LDD507 pKa = 3.23MFF509 pKa = 4.87
MM1 pKa = 7.54SATTRR6 pKa = 11.84RR7 pKa = 11.84KK8 pKa = 9.3RR9 pKa = 11.84AAPVDD14 pKa = 4.38LYY16 pKa = 11.1KK17 pKa = 10.86SCLAGGDD24 pKa = 4.48CITDD28 pKa = 3.25VKK30 pKa = 11.4NKK32 pKa = 10.28FEE34 pKa = 4.24NTTLADD40 pKa = 3.09ILLKK44 pKa = 10.81VFGSIVYY51 pKa = 10.18FGGLGIGSGKK61 pKa = 10.63GSGGSLGYY69 pKa = 10.1RR70 pKa = 11.84PLGSEE75 pKa = 3.82PTTGRR80 pKa = 11.84VAPTAPVRR88 pKa = 11.84PSVIVDD94 pKa = 3.32PLVPPDD100 pKa = 4.09VITVEE105 pKa = 4.34PSAPSIIPLAEE116 pKa = 3.89GRR118 pKa = 11.84PNIDD122 pKa = 2.96FAAPDD127 pKa = 3.92GGPGLGAEE135 pKa = 4.46EE136 pKa = 4.36IEE138 pKa = 5.03LYY140 pKa = 9.76TIKK143 pKa = 10.12TPTTDD148 pKa = 2.55IGGVGSEE155 pKa = 4.0PTVISSEE162 pKa = 4.14EE163 pKa = 3.99GATAILPVDD172 pKa = 5.13PIPEE176 pKa = 3.87RR177 pKa = 11.84PAQVFYY183 pKa = 11.21DD184 pKa = 4.41PSAPSQYY191 pKa = 10.45EE192 pKa = 3.87LNIFAAHH199 pKa = 7.36PSTSSDD205 pKa = 2.67INVFVDD211 pKa = 3.33NSFSGKK217 pKa = 9.66VVGTFEE223 pKa = 4.86EE224 pKa = 4.74IPLEE228 pKa = 4.2KK229 pKa = 10.67YY230 pKa = 10.39NYY232 pKa = 8.11STFEE236 pKa = 3.81IEE238 pKa = 4.53EE239 pKa = 4.6PPSTSTPTRR248 pKa = 11.84SLEE251 pKa = 4.13RR252 pKa = 11.84VATRR256 pKa = 11.84AKK258 pKa = 10.46ALYY261 pKa = 10.44NKK263 pKa = 7.67FTRR266 pKa = 11.84QVSVKK271 pKa = 10.46DD272 pKa = 3.88PNFLLQPSRR281 pKa = 11.84LVQFEE286 pKa = 4.12FDD288 pKa = 3.54NPAFDD293 pKa = 5.43RR294 pKa = 11.84DD295 pKa = 3.46VTLQFEE301 pKa = 4.27QDD303 pKa = 3.75VQQVTAAPNPDD314 pKa = 3.59FADD317 pKa = 4.37VIQLGRR323 pKa = 11.84PILSQTEE330 pKa = 4.04EE331 pKa = 4.17GVVRR335 pKa = 11.84VSRR338 pKa = 11.84LGKK341 pKa = 9.83VGTVATRR348 pKa = 11.84SGRR351 pKa = 11.84VIGQPVHH358 pKa = 6.04YY359 pKa = 9.4FQDD362 pKa = 3.51LSVIEE367 pKa = 4.36PEE369 pKa = 4.28SIEE372 pKa = 3.74LHH374 pKa = 6.18GFAEE378 pKa = 4.75HH379 pKa = 6.97SNQTTIVDD387 pKa = 4.24DD388 pKa = 4.53LLSSSTFQNPVFDD401 pKa = 4.46SNAVFNEE408 pKa = 3.5DD409 pKa = 4.76LLLDD413 pKa = 4.65EE414 pKa = 4.87YY415 pKa = 11.86NEE417 pKa = 4.8DD418 pKa = 3.57FGNTNLVVATTDD430 pKa = 3.22EE431 pKa = 4.4AEE433 pKa = 4.26EE434 pKa = 4.33TFSFPILAPSPSVKK448 pKa = 10.3VFVTDD453 pKa = 3.1VGNIFVTSSKK463 pKa = 10.16DD464 pKa = 3.14AHH466 pKa = 5.57STITVLNTDD475 pKa = 5.08LVPLTPSIITSSYY488 pKa = 9.36NNFYY492 pKa = 10.92LEE494 pKa = 3.96PFYY497 pKa = 11.28VPRR500 pKa = 11.84KK501 pKa = 8.28KK502 pKa = 10.14RR503 pKa = 11.84RR504 pKa = 11.84RR505 pKa = 11.84LDD507 pKa = 3.23MFF509 pKa = 4.87
Molecular weight: 55.25 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0N9E219|A0A0N9E219_9PAPI Protein E7 OS=Human papillomavirus 199 OX=1545700 GN=E7 PE=3 SV=1
MM1 pKa = 7.11EE2 pKa = 4.29TQEE5 pKa = 4.08TLTARR10 pKa = 11.84FTALQEE16 pKa = 3.99IQLTLIEE23 pKa = 4.74KK24 pKa = 10.53NSTDD28 pKa = 4.02LEE30 pKa = 4.18DD31 pKa = 6.21HH32 pKa = 5.53ITFWNAVRR40 pKa = 11.84KK41 pKa = 9.3EE42 pKa = 3.89NAIAYY47 pKa = 7.21YY48 pKa = 9.77SRR50 pKa = 11.84KK51 pKa = 9.16EE52 pKa = 3.91KK53 pKa = 10.17IYY55 pKa = 10.88KK56 pKa = 10.33LGLQPMPALAVSQYY70 pKa = 9.74KK71 pKa = 10.36AKK73 pKa = 10.09EE74 pKa = 3.97AIGIEE79 pKa = 4.05LLLRR83 pKa = 11.84SLKK86 pKa = 10.42NSQFGSEE93 pKa = 3.88RR94 pKa = 11.84WTLADD99 pKa = 4.21CSAEE103 pKa = 4.07LLHH106 pKa = 6.45TPPKK110 pKa = 10.81NCFKK114 pKa = 10.72KK115 pKa = 10.22NPYY118 pKa = 9.92SVTVLFDD125 pKa = 3.49NDD127 pKa = 3.15EE128 pKa = 4.26NKK130 pKa = 10.52AYY132 pKa = 9.49PYY134 pKa = 9.53TCWDD138 pKa = 3.63HH139 pKa = 8.03IYY141 pKa = 10.93YY142 pKa = 9.91QDD144 pKa = 5.29EE145 pKa = 3.98QNQWQKK151 pKa = 10.63VQGLVDD157 pKa = 4.07ANGMFYY163 pKa = 10.53KK164 pKa = 10.4EE165 pKa = 3.89KK166 pKa = 10.14TGDD169 pKa = 2.83IVYY172 pKa = 8.52FHH174 pKa = 6.65VFAPDD179 pKa = 3.03AEE181 pKa = 4.86TYY183 pKa = 11.2GDD185 pKa = 3.53TGEE188 pKa = 3.86WTVRR192 pKa = 11.84FKK194 pKa = 10.76NTTIFASIASSSRR207 pKa = 11.84SVSGPSEE214 pKa = 4.03QARR217 pKa = 11.84KK218 pKa = 7.7PTAHH222 pKa = 6.97SSPTPKK228 pKa = 9.89TPRR231 pKa = 11.84KK232 pKa = 8.7RR233 pKa = 11.84QRR235 pKa = 11.84EE236 pKa = 4.23TEE238 pKa = 3.99EE239 pKa = 4.11DD240 pKa = 4.1TEE242 pKa = 4.6SHH244 pKa = 6.41SPTSTSRR251 pKa = 11.84GFRR254 pKa = 11.84LRR256 pKa = 11.84RR257 pKa = 11.84GRR259 pKa = 11.84GEE261 pKa = 3.91QQGEE265 pKa = 4.58SATGPTPQRR274 pKa = 11.84RR275 pKa = 11.84RR276 pKa = 11.84RR277 pKa = 11.84TTAYY281 pKa = 9.2STVQSAPTAGEE292 pKa = 3.86VGRR295 pKa = 11.84DD296 pKa = 3.32TRR298 pKa = 11.84SVPRR302 pKa = 11.84HH303 pKa = 4.76GLDD306 pKa = 2.89RR307 pKa = 11.84LRR309 pKa = 11.84RR310 pKa = 11.84LQAEE314 pKa = 3.87ARR316 pKa = 11.84DD317 pKa = 3.89PFLVLLKK324 pKa = 10.78GPPNCLKK331 pKa = 10.21CWRR334 pKa = 11.84YY335 pKa = 9.62RR336 pKa = 11.84CKK338 pKa = 10.53QKK340 pKa = 11.15NNAMFLAVSSVFRR353 pKa = 11.84WIGDD357 pKa = 3.33EE358 pKa = 4.31SEE360 pKa = 4.03SHH362 pKa = 6.61ARR364 pKa = 11.84LLLAFSDD371 pKa = 3.23NRR373 pKa = 11.84QRR375 pKa = 11.84QTFLDD380 pKa = 3.54NVTFPKK386 pKa = 9.88GTTYY390 pKa = 11.32SLGNIDD396 pKa = 4.41SLL398 pKa = 4.54
MM1 pKa = 7.11EE2 pKa = 4.29TQEE5 pKa = 4.08TLTARR10 pKa = 11.84FTALQEE16 pKa = 3.99IQLTLIEE23 pKa = 4.74KK24 pKa = 10.53NSTDD28 pKa = 4.02LEE30 pKa = 4.18DD31 pKa = 6.21HH32 pKa = 5.53ITFWNAVRR40 pKa = 11.84KK41 pKa = 9.3EE42 pKa = 3.89NAIAYY47 pKa = 7.21YY48 pKa = 9.77SRR50 pKa = 11.84KK51 pKa = 9.16EE52 pKa = 3.91KK53 pKa = 10.17IYY55 pKa = 10.88KK56 pKa = 10.33LGLQPMPALAVSQYY70 pKa = 9.74KK71 pKa = 10.36AKK73 pKa = 10.09EE74 pKa = 3.97AIGIEE79 pKa = 4.05LLLRR83 pKa = 11.84SLKK86 pKa = 10.42NSQFGSEE93 pKa = 3.88RR94 pKa = 11.84WTLADD99 pKa = 4.21CSAEE103 pKa = 4.07LLHH106 pKa = 6.45TPPKK110 pKa = 10.81NCFKK114 pKa = 10.72KK115 pKa = 10.22NPYY118 pKa = 9.92SVTVLFDD125 pKa = 3.49NDD127 pKa = 3.15EE128 pKa = 4.26NKK130 pKa = 10.52AYY132 pKa = 9.49PYY134 pKa = 9.53TCWDD138 pKa = 3.63HH139 pKa = 8.03IYY141 pKa = 10.93YY142 pKa = 9.91QDD144 pKa = 5.29EE145 pKa = 3.98QNQWQKK151 pKa = 10.63VQGLVDD157 pKa = 4.07ANGMFYY163 pKa = 10.53KK164 pKa = 10.4EE165 pKa = 3.89KK166 pKa = 10.14TGDD169 pKa = 2.83IVYY172 pKa = 8.52FHH174 pKa = 6.65VFAPDD179 pKa = 3.03AEE181 pKa = 4.86TYY183 pKa = 11.2GDD185 pKa = 3.53TGEE188 pKa = 3.86WTVRR192 pKa = 11.84FKK194 pKa = 10.76NTTIFASIASSSRR207 pKa = 11.84SVSGPSEE214 pKa = 4.03QARR217 pKa = 11.84KK218 pKa = 7.7PTAHH222 pKa = 6.97SSPTPKK228 pKa = 9.89TPRR231 pKa = 11.84KK232 pKa = 8.7RR233 pKa = 11.84QRR235 pKa = 11.84EE236 pKa = 4.23TEE238 pKa = 3.99EE239 pKa = 4.11DD240 pKa = 4.1TEE242 pKa = 4.6SHH244 pKa = 6.41SPTSTSRR251 pKa = 11.84GFRR254 pKa = 11.84LRR256 pKa = 11.84RR257 pKa = 11.84GRR259 pKa = 11.84GEE261 pKa = 3.91QQGEE265 pKa = 4.58SATGPTPQRR274 pKa = 11.84RR275 pKa = 11.84RR276 pKa = 11.84RR277 pKa = 11.84TTAYY281 pKa = 9.2STVQSAPTAGEE292 pKa = 3.86VGRR295 pKa = 11.84DD296 pKa = 3.32TRR298 pKa = 11.84SVPRR302 pKa = 11.84HH303 pKa = 4.76GLDD306 pKa = 2.89RR307 pKa = 11.84LRR309 pKa = 11.84RR310 pKa = 11.84LQAEE314 pKa = 3.87ARR316 pKa = 11.84DD317 pKa = 3.89PFLVLLKK324 pKa = 10.78GPPNCLKK331 pKa = 10.21CWRR334 pKa = 11.84YY335 pKa = 9.62RR336 pKa = 11.84CKK338 pKa = 10.53QKK340 pKa = 11.15NNAMFLAVSSVFRR353 pKa = 11.84WIGDD357 pKa = 3.33EE358 pKa = 4.31SEE360 pKa = 4.03SHH362 pKa = 6.61ARR364 pKa = 11.84LLLAFSDD371 pKa = 3.23NRR373 pKa = 11.84QRR375 pKa = 11.84QTFLDD380 pKa = 3.54NVTFPKK386 pKa = 9.88GTTYY390 pKa = 11.32SLGNIDD396 pKa = 4.41SLL398 pKa = 4.54
Molecular weight: 45.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2248 |
97 |
598 |
374.7 |
42.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.361 ± 0.321 | 2.447 ± 0.877 |
6.272 ± 0.467 | 6.094 ± 0.542 |
5.605 ± 0.485 | 4.938 ± 0.859 |
1.824 ± 0.442 | 4.893 ± 0.424 |
5.472 ± 0.643 | 8.763 ± 0.709 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.69 ± 0.408 | 5.249 ± 0.593 |
5.694 ± 1.049 | 4.004 ± 0.313 |
5.516 ± 0.642 | 7.562 ± 0.421 |
6.673 ± 0.647 | 6.228 ± 0.724 |
1.201 ± 0.281 | 3.514 ± 0.262 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
