
Azorhizobium caulinodans (strain ATCC 43989 / DSM 5975 / JCM 20966 / NBRC 14845 / NCIMB 13405 / ORS 571)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Xanthobacteraceae; Azorhizobium; Azorhizobium caulinodans
Average proteome isoelectric point is 6.78
Get precalculated fractions of proteins
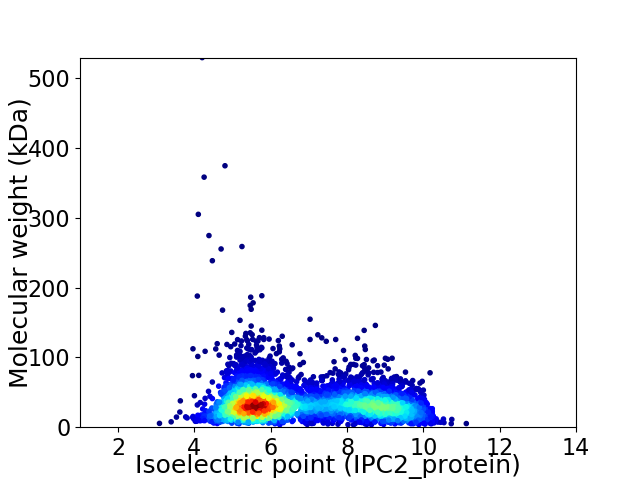
Virtual 2D-PAGE plot for 4708 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A8IDJ4|A8IDJ4_AZOC5 Peptidase OS=Azorhizobium caulinodans (strain ATCC 43989 / DSM 5975 / JCM 20966 / NBRC 14845 / NCIMB 13405 / ORS 571) OX=438753 GN=AZC_2950 PE=3 SV=1
MM1 pKa = 7.8AKK3 pKa = 7.34TTTITWSWGTNSVIAFDD20 pKa = 4.23PASDD24 pKa = 3.37ILDD27 pKa = 4.09FGWFQAGQFTVSEE40 pKa = 4.61VNGSVVISIPTNNQTYY56 pKa = 8.17TLTGVTLSEE65 pKa = 4.37LSLSNILAKK74 pKa = 10.76DD75 pKa = 3.42SGAVTEE81 pKa = 4.38WTTALGGASAGSGGSSGGATSGGATSGGTSTGGSAGGSPASGTTDD126 pKa = 2.81GGTSGSSGGAGHH138 pKa = 6.78VGDD141 pKa = 4.79GLATVTTISWAWGTHH156 pKa = 3.84TALNFDD162 pKa = 3.9TALDD166 pKa = 3.72KK167 pKa = 11.51LDD169 pKa = 4.68FGWLSADD176 pKa = 3.03NFTVSEE182 pKa = 4.47VNGSVVIAIPSNSQTYY198 pKa = 8.14TLTGVTLSEE207 pKa = 4.23LSLANIVAKK216 pKa = 10.64DD217 pKa = 3.79GSALAEE223 pKa = 4.03WTTALSGHH231 pKa = 6.28SGGSSGTSGGSTGGSTSGGGSSSAGGSTSTGGGSTGGGSTGGSSTGGSTGGTSGGGTSGGDD292 pKa = 2.96AALYY296 pKa = 10.41AEE298 pKa = 4.91AWSASKK304 pKa = 10.66VYY306 pKa = 10.63QGGDD310 pKa = 2.98HH311 pKa = 6.81AAVGNNVYY319 pKa = 9.38EE320 pKa = 4.29AHH322 pKa = 7.05WWTQGTDD329 pKa = 3.3PSTHH333 pKa = 6.22NGGDD337 pKa = 3.2GSGQVWTLVGHH348 pKa = 7.46LDD350 pKa = 3.54TTTVVPNAPADD361 pKa = 3.77LFATNVSDD369 pKa = 4.32TSALLVWDD377 pKa = 3.76KK378 pKa = 11.58AVINGAGTVSAYY390 pKa = 9.99EE391 pKa = 3.88IYY393 pKa = 10.85EE394 pKa = 4.13NGTLVGTTSGTSYY407 pKa = 11.2KK408 pKa = 9.5VTGLGASTAYY418 pKa = 10.51SFTVVAVDD426 pKa = 3.7EE427 pKa = 5.24AGHH430 pKa = 6.03SPAAVPISVTTDD442 pKa = 3.36PVHH445 pKa = 6.45TSTLEE450 pKa = 3.9QTYY453 pKa = 10.58SPYY456 pKa = 10.25IDD458 pKa = 3.85MSLSTSQDD466 pKa = 3.26LLSIAKK472 pKa = 10.2ASGVTDD478 pKa = 4.04FTLAFVLSSGTDD490 pKa = 3.21TLGWGGVGTLGNDD503 pKa = 3.65TLPSGSSIHH512 pKa = 6.58DD513 pKa = 3.17QVAAVQAIGGDD524 pKa = 3.35ITISFGGANGQEE536 pKa = 3.95AALTFSSATKK546 pKa = 9.2LTAAYY551 pKa = 10.1QSVLDD556 pKa = 4.11TYY558 pKa = 10.73HH559 pKa = 6.18VNKK562 pKa = 9.86IDD564 pKa = 3.68FDD566 pKa = 4.11IEE568 pKa = 3.78GGAIANTSANHH579 pKa = 6.26LRR581 pKa = 11.84DD582 pKa = 3.38QALVALEE589 pKa = 4.25AANPDD594 pKa = 3.66LKK596 pKa = 11.33VSFTLPVLPTGLTNDD611 pKa = 3.93GLNLLKK617 pKa = 10.46QALTDD622 pKa = 3.8GVHH625 pKa = 6.56IDD627 pKa = 3.86TVNIMAMDD635 pKa = 4.02YY636 pKa = 10.56GASVDD641 pKa = 3.93SGDD644 pKa = 3.95MGTDD648 pKa = 4.03AISAAQATLAQMHH661 pKa = 5.97TLGLDD666 pKa = 3.01AKK668 pKa = 10.64LGVTVMVGMNDD679 pKa = 3.23VQSEE683 pKa = 4.72VFTLSDD689 pKa = 3.38AQQLLDD695 pKa = 3.66YY696 pKa = 10.85AQGNEE701 pKa = 4.6DD702 pKa = 3.95ISGLSIWSVGRR713 pKa = 11.84DD714 pKa = 3.13NGSTVGTVSPLGSGIQQTAYY734 pKa = 9.67EE735 pKa = 4.11FSHH738 pKa = 7.1IFGHH742 pKa = 6.8II743 pKa = 3.2
MM1 pKa = 7.8AKK3 pKa = 7.34TTTITWSWGTNSVIAFDD20 pKa = 4.23PASDD24 pKa = 3.37ILDD27 pKa = 4.09FGWFQAGQFTVSEE40 pKa = 4.61VNGSVVISIPTNNQTYY56 pKa = 8.17TLTGVTLSEE65 pKa = 4.37LSLSNILAKK74 pKa = 10.76DD75 pKa = 3.42SGAVTEE81 pKa = 4.38WTTALGGASAGSGGSSGGATSGGATSGGTSTGGSAGGSPASGTTDD126 pKa = 2.81GGTSGSSGGAGHH138 pKa = 6.78VGDD141 pKa = 4.79GLATVTTISWAWGTHH156 pKa = 3.84TALNFDD162 pKa = 3.9TALDD166 pKa = 3.72KK167 pKa = 11.51LDD169 pKa = 4.68FGWLSADD176 pKa = 3.03NFTVSEE182 pKa = 4.47VNGSVVIAIPSNSQTYY198 pKa = 8.14TLTGVTLSEE207 pKa = 4.23LSLANIVAKK216 pKa = 10.64DD217 pKa = 3.79GSALAEE223 pKa = 4.03WTTALSGHH231 pKa = 6.28SGGSSGTSGGSTGGSTSGGGSSSAGGSTSTGGGSTGGGSTGGSSTGGSTGGTSGGGTSGGDD292 pKa = 2.96AALYY296 pKa = 10.41AEE298 pKa = 4.91AWSASKK304 pKa = 10.66VYY306 pKa = 10.63QGGDD310 pKa = 2.98HH311 pKa = 6.81AAVGNNVYY319 pKa = 9.38EE320 pKa = 4.29AHH322 pKa = 7.05WWTQGTDD329 pKa = 3.3PSTHH333 pKa = 6.22NGGDD337 pKa = 3.2GSGQVWTLVGHH348 pKa = 7.46LDD350 pKa = 3.54TTTVVPNAPADD361 pKa = 3.77LFATNVSDD369 pKa = 4.32TSALLVWDD377 pKa = 3.76KK378 pKa = 11.58AVINGAGTVSAYY390 pKa = 9.99EE391 pKa = 3.88IYY393 pKa = 10.85EE394 pKa = 4.13NGTLVGTTSGTSYY407 pKa = 11.2KK408 pKa = 9.5VTGLGASTAYY418 pKa = 10.51SFTVVAVDD426 pKa = 3.7EE427 pKa = 5.24AGHH430 pKa = 6.03SPAAVPISVTTDD442 pKa = 3.36PVHH445 pKa = 6.45TSTLEE450 pKa = 3.9QTYY453 pKa = 10.58SPYY456 pKa = 10.25IDD458 pKa = 3.85MSLSTSQDD466 pKa = 3.26LLSIAKK472 pKa = 10.2ASGVTDD478 pKa = 4.04FTLAFVLSSGTDD490 pKa = 3.21TLGWGGVGTLGNDD503 pKa = 3.65TLPSGSSIHH512 pKa = 6.58DD513 pKa = 3.17QVAAVQAIGGDD524 pKa = 3.35ITISFGGANGQEE536 pKa = 3.95AALTFSSATKK546 pKa = 9.2LTAAYY551 pKa = 10.1QSVLDD556 pKa = 4.11TYY558 pKa = 10.73HH559 pKa = 6.18VNKK562 pKa = 9.86IDD564 pKa = 3.68FDD566 pKa = 4.11IEE568 pKa = 3.78GGAIANTSANHH579 pKa = 6.26LRR581 pKa = 11.84DD582 pKa = 3.38QALVALEE589 pKa = 4.25AANPDD594 pKa = 3.66LKK596 pKa = 11.33VSFTLPVLPTGLTNDD611 pKa = 3.93GLNLLKK617 pKa = 10.46QALTDD622 pKa = 3.8GVHH625 pKa = 6.56IDD627 pKa = 3.86TVNIMAMDD635 pKa = 4.02YY636 pKa = 10.56GASVDD641 pKa = 3.93SGDD644 pKa = 3.95MGTDD648 pKa = 4.03AISAAQATLAQMHH661 pKa = 5.97TLGLDD666 pKa = 3.01AKK668 pKa = 10.64LGVTVMVGMNDD679 pKa = 3.23VQSEE683 pKa = 4.72VFTLSDD689 pKa = 3.38AQQLLDD695 pKa = 3.66YY696 pKa = 10.85AQGNEE701 pKa = 4.6DD702 pKa = 3.95ISGLSIWSVGRR713 pKa = 11.84DD714 pKa = 3.13NGSTVGTVSPLGSGIQQTAYY734 pKa = 9.67EE735 pKa = 4.11FSHH738 pKa = 7.1IFGHH742 pKa = 6.8II743 pKa = 3.2
Molecular weight: 73.86 kDa
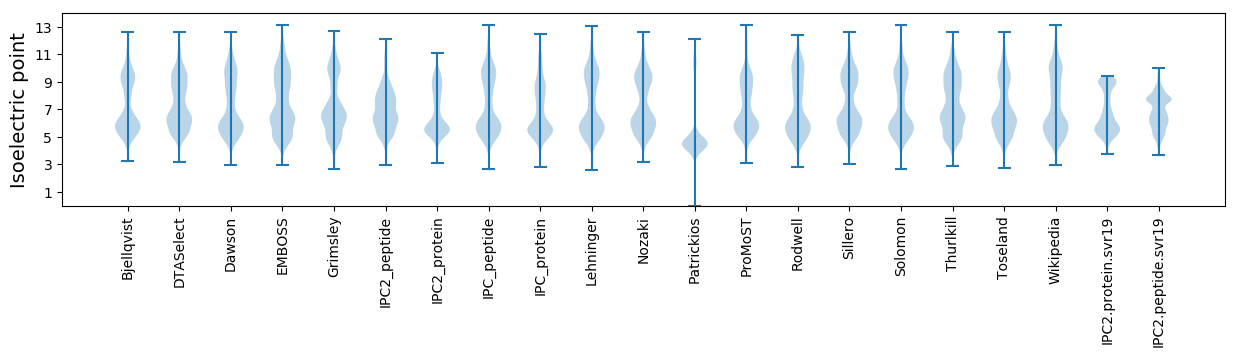
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A8IM23|A8IM23_AZOC5 Formyl-CoA:oxalate CoA-transferase OS=Azorhizobium caulinodans (strain ATCC 43989 / DSM 5975 / JCM 20966 / NBRC 14845 / NCIMB 13405 / ORS 571) OX=438753 GN=frc PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.27NGRR28 pKa = 11.84KK29 pKa = 9.24IIAARR34 pKa = 11.84RR35 pKa = 11.84AHH37 pKa = 5.12GRR39 pKa = 11.84KK40 pKa = 9.3RR41 pKa = 11.84LSAA44 pKa = 4.01
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.27QPSKK9 pKa = 9.73LVRR12 pKa = 11.84KK13 pKa = 9.15RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.27NGRR28 pKa = 11.84KK29 pKa = 9.24IIAARR34 pKa = 11.84RR35 pKa = 11.84AHH37 pKa = 5.12GRR39 pKa = 11.84KK40 pKa = 9.3RR41 pKa = 11.84LSAA44 pKa = 4.01
Molecular weight: 5.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1602785 |
30 |
5585 |
340.4 |
36.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.567 ± 0.053 | 0.814 ± 0.011 |
5.277 ± 0.03 | 5.336 ± 0.045 |
3.626 ± 0.022 | 8.886 ± 0.071 |
1.949 ± 0.02 | 4.665 ± 0.027 |
2.929 ± 0.035 | 10.449 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.36 ± 0.017 | 2.35 ± 0.026 |
5.664 ± 0.039 | 2.978 ± 0.02 |
7.229 ± 0.052 | 5.288 ± 0.046 |
5.492 ± 0.061 | 7.796 ± 0.029 |
1.246 ± 0.015 | 2.1 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
