
Avon-Heathcote Estuary associated circular virus 23
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 8.34
Get precalculated fractions of proteins
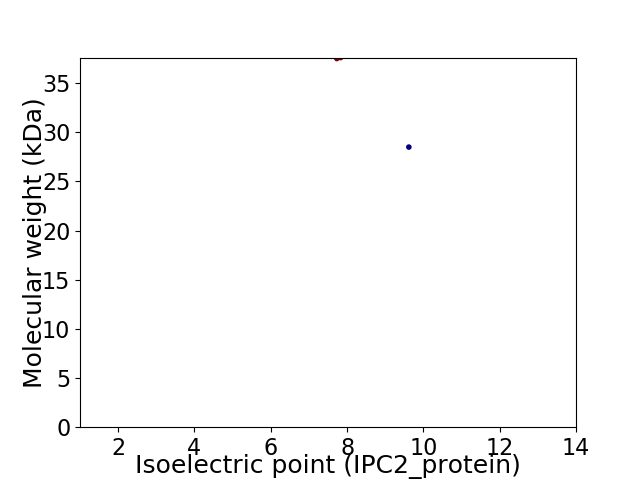
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5IMK7|A0A0C5IMK7_9CIRC Putative capsid protein OS=Avon-Heathcote Estuary associated circular virus 23 OX=1618247 PE=4 SV=1
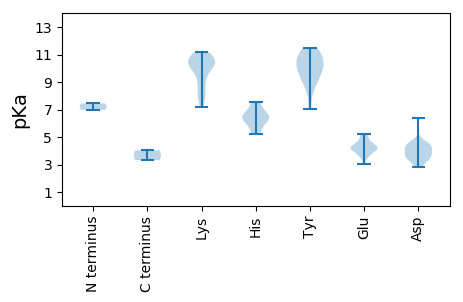
MM1 pKa = 7.44TARR4 pKa = 11.84NWCFTLNNPTEE15 pKa = 4.84HH16 pKa = 7.21IDD18 pKa = 3.77FKK20 pKa = 10.74SEE22 pKa = 3.05KK23 pKa = 10.21WMDD26 pKa = 3.75LKK28 pKa = 11.11LAIYY32 pKa = 9.62MMEE35 pKa = 4.45KK36 pKa = 10.78GEE38 pKa = 4.22TGTPHH43 pKa = 5.24FQGYY47 pKa = 10.36LEE49 pKa = 4.16MKK51 pKa = 9.25EE52 pKa = 4.08PQRR55 pKa = 11.84MSFFKK60 pKa = 10.86KK61 pKa = 10.25LMPRR65 pKa = 11.84AHH67 pKa = 6.21LEE69 pKa = 3.56KK70 pKa = 10.52RR71 pKa = 11.84RR72 pKa = 11.84GSKK75 pKa = 9.13QQAINYY81 pKa = 7.04VLKK84 pKa = 10.32TMDD87 pKa = 4.36PEE89 pKa = 4.5CSQWEE94 pKa = 3.91ITMTGEE100 pKa = 3.93QPIITGLPVAIHH112 pKa = 6.5CKK114 pKa = 8.9GTPHH118 pKa = 7.57DD119 pKa = 4.53LLKK122 pKa = 10.94SCEE125 pKa = 4.16PKK127 pKa = 10.45KK128 pKa = 10.6SLKK131 pKa = 10.05EE132 pKa = 3.61RR133 pKa = 11.84LEE135 pKa = 4.04DD136 pKa = 3.45VKK138 pKa = 11.16EE139 pKa = 4.45LIRR142 pKa = 11.84SGYY145 pKa = 9.76NDD147 pKa = 3.52EE148 pKa = 4.99QIMEE152 pKa = 4.06SHH154 pKa = 6.56YY155 pKa = 11.24DD156 pKa = 3.3LFVRR160 pKa = 11.84YY161 pKa = 8.97HH162 pKa = 6.46RR163 pKa = 11.84AFALTRR169 pKa = 11.84RR170 pKa = 11.84IMSSPRR176 pKa = 11.84DD177 pKa = 3.55HH178 pKa = 6.55EE179 pKa = 4.34MKK181 pKa = 10.77VIVCQGPTGTGKK193 pKa = 10.4SRR195 pKa = 11.84WAKK198 pKa = 7.72EE199 pKa = 4.28TYY201 pKa = 9.92PNAYY205 pKa = 8.08WKK207 pKa = 10.03QRR209 pKa = 11.84SQWWCGYY216 pKa = 9.12EE217 pKa = 4.11GQDD220 pKa = 4.43TIVLDD225 pKa = 4.25EE226 pKa = 4.62FYY228 pKa = 11.22GWLQYY233 pKa = 11.47DD234 pKa = 4.37LLLRR238 pKa = 11.84LCDD241 pKa = 4.16RR242 pKa = 11.84YY243 pKa = 10.26PLYY246 pKa = 11.16LEE248 pKa = 4.36TKK250 pKa = 9.84GGQVNCKK257 pKa = 9.96AKK259 pKa = 10.17TIVITTNATPEE270 pKa = 3.79SWYY273 pKa = 9.1KK274 pKa = 7.79TEE276 pKa = 5.02KK277 pKa = 10.73YY278 pKa = 9.44FLSFIRR284 pKa = 11.84RR285 pKa = 11.84VSEE288 pKa = 3.51WRR290 pKa = 11.84VFPIWGEE297 pKa = 3.81MEE299 pKa = 4.27TYY301 pKa = 10.51VLYY304 pKa = 11.03SEE306 pKa = 5.25AVSKK310 pKa = 10.46FFLNTEE316 pKa = 4.03
MM1 pKa = 7.44TARR4 pKa = 11.84NWCFTLNNPTEE15 pKa = 4.84HH16 pKa = 7.21IDD18 pKa = 3.77FKK20 pKa = 10.74SEE22 pKa = 3.05KK23 pKa = 10.21WMDD26 pKa = 3.75LKK28 pKa = 11.11LAIYY32 pKa = 9.62MMEE35 pKa = 4.45KK36 pKa = 10.78GEE38 pKa = 4.22TGTPHH43 pKa = 5.24FQGYY47 pKa = 10.36LEE49 pKa = 4.16MKK51 pKa = 9.25EE52 pKa = 4.08PQRR55 pKa = 11.84MSFFKK60 pKa = 10.86KK61 pKa = 10.25LMPRR65 pKa = 11.84AHH67 pKa = 6.21LEE69 pKa = 3.56KK70 pKa = 10.52RR71 pKa = 11.84RR72 pKa = 11.84GSKK75 pKa = 9.13QQAINYY81 pKa = 7.04VLKK84 pKa = 10.32TMDD87 pKa = 4.36PEE89 pKa = 4.5CSQWEE94 pKa = 3.91ITMTGEE100 pKa = 3.93QPIITGLPVAIHH112 pKa = 6.5CKK114 pKa = 8.9GTPHH118 pKa = 7.57DD119 pKa = 4.53LLKK122 pKa = 10.94SCEE125 pKa = 4.16PKK127 pKa = 10.45KK128 pKa = 10.6SLKK131 pKa = 10.05EE132 pKa = 3.61RR133 pKa = 11.84LEE135 pKa = 4.04DD136 pKa = 3.45VKK138 pKa = 11.16EE139 pKa = 4.45LIRR142 pKa = 11.84SGYY145 pKa = 9.76NDD147 pKa = 3.52EE148 pKa = 4.99QIMEE152 pKa = 4.06SHH154 pKa = 6.56YY155 pKa = 11.24DD156 pKa = 3.3LFVRR160 pKa = 11.84YY161 pKa = 8.97HH162 pKa = 6.46RR163 pKa = 11.84AFALTRR169 pKa = 11.84RR170 pKa = 11.84IMSSPRR176 pKa = 11.84DD177 pKa = 3.55HH178 pKa = 6.55EE179 pKa = 4.34MKK181 pKa = 10.77VIVCQGPTGTGKK193 pKa = 10.4SRR195 pKa = 11.84WAKK198 pKa = 7.72EE199 pKa = 4.28TYY201 pKa = 9.92PNAYY205 pKa = 8.08WKK207 pKa = 10.03QRR209 pKa = 11.84SQWWCGYY216 pKa = 9.12EE217 pKa = 4.11GQDD220 pKa = 4.43TIVLDD225 pKa = 4.25EE226 pKa = 4.62FYY228 pKa = 11.22GWLQYY233 pKa = 11.47DD234 pKa = 4.37LLLRR238 pKa = 11.84LCDD241 pKa = 4.16RR242 pKa = 11.84YY243 pKa = 10.26PLYY246 pKa = 11.16LEE248 pKa = 4.36TKK250 pKa = 9.84GGQVNCKK257 pKa = 9.96AKK259 pKa = 10.17TIVITTNATPEE270 pKa = 3.79SWYY273 pKa = 9.1KK274 pKa = 7.79TEE276 pKa = 5.02KK277 pKa = 10.73YY278 pKa = 9.44FLSFIRR284 pKa = 11.84RR285 pKa = 11.84VSEE288 pKa = 3.51WRR290 pKa = 11.84VFPIWGEE297 pKa = 3.81MEE299 pKa = 4.27TYY301 pKa = 10.51VLYY304 pKa = 11.03SEE306 pKa = 5.25AVSKK310 pKa = 10.46FFLNTEE316 pKa = 4.03
Molecular weight: 37.5 kDa
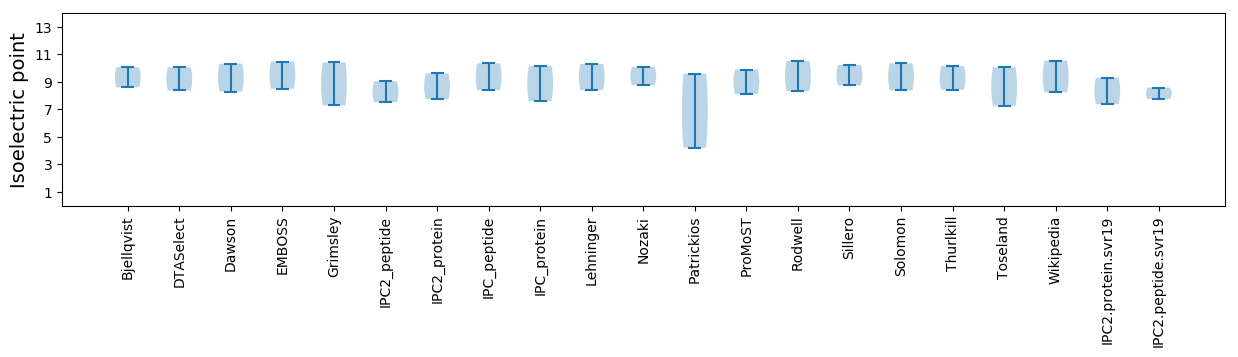
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5IMK7|A0A0C5IMK7_9CIRC Putative capsid protein OS=Avon-Heathcote Estuary associated circular virus 23 OX=1618247 PE=4 SV=1
MM1 pKa = 6.98VFARR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 8.19RR8 pKa = 11.84KK9 pKa = 7.87TYY11 pKa = 10.54AGRR14 pKa = 11.84KK15 pKa = 7.2RR16 pKa = 11.84SRR18 pKa = 11.84RR19 pKa = 11.84SFKK22 pKa = 10.16RR23 pKa = 11.84RR24 pKa = 11.84SNVKK28 pKa = 8.56RR29 pKa = 11.84RR30 pKa = 11.84MVRR33 pKa = 11.84RR34 pKa = 11.84PRR36 pKa = 11.84NIGRR40 pKa = 11.84LPGFPEE46 pKa = 4.32TYY48 pKa = 8.89KK49 pKa = 10.57TVLVYY54 pKa = 11.12NEE56 pKa = 5.2GISIDD61 pKa = 3.73AAAGGATYY69 pKa = 11.13YY70 pKa = 11.06SFNCMNLYY78 pKa = 10.29DD79 pKa = 4.5PNYY82 pKa = 9.96SGTGHH87 pKa = 6.24QPLYY91 pKa = 10.37YY92 pKa = 10.54DD93 pKa = 4.28NLTAIYY99 pKa = 10.08NRR101 pKa = 11.84WRR103 pKa = 11.84VRR105 pKa = 11.84AAYY108 pKa = 8.33ITVTILDD115 pKa = 4.13TFVSNATVDD124 pKa = 3.06AGAAYY129 pKa = 9.66RR130 pKa = 11.84AQGTGGRR137 pKa = 11.84LIVCRR142 pKa = 11.84DD143 pKa = 2.79INAADD148 pKa = 4.47NEE150 pKa = 4.67TTTNNIQEE158 pKa = 4.24EE159 pKa = 4.16RR160 pKa = 11.84SKK162 pKa = 10.92NIKK165 pKa = 8.3WRR167 pKa = 11.84YY168 pKa = 9.31FSAQTNGRR176 pKa = 11.84DD177 pKa = 3.65PKK179 pKa = 10.76LKK181 pKa = 9.93MMCIPHH187 pKa = 5.61VQLKK191 pKa = 9.08FAKK194 pKa = 10.25DD195 pKa = 3.34DD196 pKa = 3.71AQLFGSVGSGPTANCFFNIGVAGVDD221 pKa = 3.52DD222 pKa = 4.46TVNPPNVRR230 pKa = 11.84LNVRR234 pKa = 11.84ISYY237 pKa = 10.35VVDD240 pKa = 4.34FFDD243 pKa = 6.4RR244 pKa = 11.84IVDD247 pKa = 3.67QPQNN251 pKa = 3.31
MM1 pKa = 6.98VFARR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 8.19RR8 pKa = 11.84KK9 pKa = 7.87TYY11 pKa = 10.54AGRR14 pKa = 11.84KK15 pKa = 7.2RR16 pKa = 11.84SRR18 pKa = 11.84RR19 pKa = 11.84SFKK22 pKa = 10.16RR23 pKa = 11.84RR24 pKa = 11.84SNVKK28 pKa = 8.56RR29 pKa = 11.84RR30 pKa = 11.84MVRR33 pKa = 11.84RR34 pKa = 11.84PRR36 pKa = 11.84NIGRR40 pKa = 11.84LPGFPEE46 pKa = 4.32TYY48 pKa = 8.89KK49 pKa = 10.57TVLVYY54 pKa = 11.12NEE56 pKa = 5.2GISIDD61 pKa = 3.73AAAGGATYY69 pKa = 11.13YY70 pKa = 11.06SFNCMNLYY78 pKa = 10.29DD79 pKa = 4.5PNYY82 pKa = 9.96SGTGHH87 pKa = 6.24QPLYY91 pKa = 10.37YY92 pKa = 10.54DD93 pKa = 4.28NLTAIYY99 pKa = 10.08NRR101 pKa = 11.84WRR103 pKa = 11.84VRR105 pKa = 11.84AAYY108 pKa = 8.33ITVTILDD115 pKa = 4.13TFVSNATVDD124 pKa = 3.06AGAAYY129 pKa = 9.66RR130 pKa = 11.84AQGTGGRR137 pKa = 11.84LIVCRR142 pKa = 11.84DD143 pKa = 2.79INAADD148 pKa = 4.47NEE150 pKa = 4.67TTTNNIQEE158 pKa = 4.24EE159 pKa = 4.16RR160 pKa = 11.84SKK162 pKa = 10.92NIKK165 pKa = 8.3WRR167 pKa = 11.84YY168 pKa = 9.31FSAQTNGRR176 pKa = 11.84DD177 pKa = 3.65PKK179 pKa = 10.76LKK181 pKa = 9.93MMCIPHH187 pKa = 5.61VQLKK191 pKa = 9.08FAKK194 pKa = 10.25DD195 pKa = 3.34DD196 pKa = 3.71AQLFGSVGSGPTANCFFNIGVAGVDD221 pKa = 3.52DD222 pKa = 4.46TVNPPNVRR230 pKa = 11.84LNVRR234 pKa = 11.84ISYY237 pKa = 10.35VVDD240 pKa = 4.34FFDD243 pKa = 6.4RR244 pKa = 11.84IVDD247 pKa = 3.67QPQNN251 pKa = 3.31
Molecular weight: 28.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
567 |
251 |
316 |
283.5 |
33.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.82 ± 1.576 | 2.116 ± 0.324 |
4.762 ± 0.752 | 5.82 ± 2.37 |
4.409 ± 0.23 | 6.173 ± 0.618 |
1.764 ± 0.599 | 5.291 ± 0.177 |
6.702 ± 1.189 | 6.526 ± 1.327 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.175 ± 0.732 | 5.291 ± 1.904 |
4.586 ± 0.126 | 3.704 ± 0.32 |
8.113 ± 1.637 | 5.115 ± 0.207 |
7.055 ± 0.174 | 5.82 ± 1.33 |
2.293 ± 0.926 | 5.467 ± 0.068 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
