
Trichomonas vaginalis virus 1 (TVV1)
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; Trichomonasvirus
Average proteome isoelectric point is 6.8
Get precalculated fractions of proteins
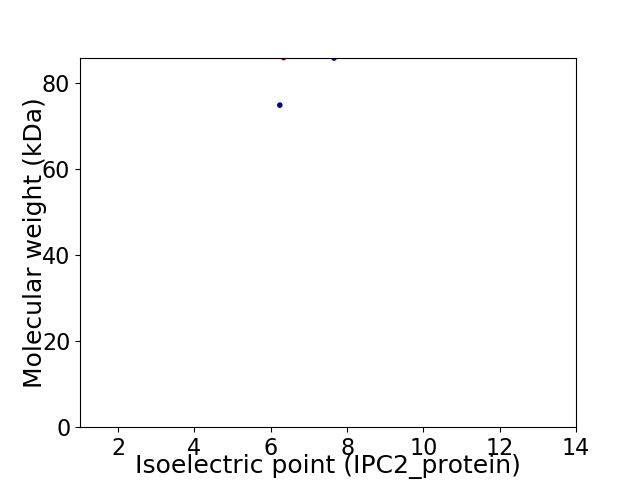
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q90155|Q90155_TTV1 RNA-directed RNA polymerase (Fragment) OS=Trichomonas vaginalis virus 1 OX=674953 GN=pol PE=3 SV=1
MM1 pKa = 7.19EE2 pKa = 5.64ASANGLSHH10 pKa = 7.93DD11 pKa = 4.42DD12 pKa = 4.09NANKK16 pKa = 9.65SQNVGPSTLPRR27 pKa = 11.84SDD29 pKa = 3.14KK30 pKa = 11.09QGGEE34 pKa = 3.64KK35 pKa = 10.34HH36 pKa = 6.28EE37 pKa = 4.3NSFNSFSNDD46 pKa = 2.5FFFNFLRR53 pKa = 11.84MSTNTHH59 pKa = 6.17ISDD62 pKa = 3.74SPGVSFVAKK71 pKa = 10.42DD72 pKa = 3.41GTPYY76 pKa = 10.95SSLTIPSGVGRR87 pKa = 11.84LTHH90 pKa = 6.16NVVASAVQLNITASNTLEE108 pKa = 3.65VDD110 pKa = 3.38YY111 pKa = 11.5GFGQDD116 pKa = 3.26VSRR119 pKa = 11.84TTGTIPIPIFDD130 pKa = 3.77GEE132 pKa = 4.61KK133 pKa = 9.59YY134 pKa = 10.29KK135 pKa = 10.09EE136 pKa = 4.0TARR139 pKa = 11.84ALAAIFSKK147 pKa = 10.62KK148 pKa = 10.06GMSVDD153 pKa = 3.52VTSQTVQEE161 pKa = 4.23TLKK164 pKa = 11.21NSDD167 pKa = 3.33LTIATVAAGYY177 pKa = 7.73YY178 pKa = 7.0TALAARR184 pKa = 11.84HH185 pKa = 5.96EE186 pKa = 4.28LTKK189 pKa = 10.79DD190 pKa = 3.28VSEE193 pKa = 4.52AAHH196 pKa = 6.69TIPFVTALSDD206 pKa = 3.54TFSAALNAQRR216 pKa = 11.84TSHH219 pKa = 6.43VISSCLRR226 pKa = 11.84CPNSRR231 pKa = 11.84NAQRR235 pKa = 11.84DD236 pKa = 3.62IVIGTVLWNNVFVEE250 pKa = 4.4SLSEE254 pKa = 3.98HH255 pKa = 5.97NMAVPNPNDD264 pKa = 2.91ISFFIPNKK272 pKa = 9.99ALSSSWWCAIWLLNAFLHH290 pKa = 6.5SFIAPTRR297 pKa = 11.84IHH299 pKa = 6.94IFITQGEE306 pKa = 4.94TYY308 pKa = 10.07HH309 pKa = 6.92LAPFTDD315 pKa = 3.16SDD317 pKa = 3.84VYY319 pKa = 10.42EE320 pKa = 4.07AVRR323 pKa = 11.84FLLAMSKK330 pKa = 10.16SSRR333 pKa = 11.84PMPEE337 pKa = 3.86SVEE340 pKa = 4.31SMLYY344 pKa = 10.39AYY346 pKa = 7.32GTQMIIQPHH355 pKa = 4.82SLYY358 pKa = 10.63TEE360 pKa = 4.42GGLIRR365 pKa = 11.84RR366 pKa = 11.84MIFTVPHH373 pKa = 6.59LPAHH377 pKa = 6.9GYY379 pKa = 9.65FVTNSEE385 pKa = 4.1FSRR388 pKa = 11.84YY389 pKa = 7.39MNIAVPDD396 pKa = 4.1DD397 pKa = 3.67PRR399 pKa = 11.84SAKK402 pKa = 10.69DD403 pKa = 3.21FVIGAGTGLLQIVLAYY419 pKa = 9.72QAAFSCAGPIALHH432 pKa = 5.63WHH434 pKa = 6.29ANDD437 pKa = 5.58AISQGMDD444 pKa = 2.8RR445 pKa = 11.84VASIYY450 pKa = 10.99LEE452 pKa = 3.79GRR454 pKa = 11.84YY455 pKa = 7.58FTIPMAVNVATNVAQYY471 pKa = 7.91TTMVRR476 pKa = 11.84ADD478 pKa = 3.19PEE480 pKa = 4.18YY481 pKa = 10.68RR482 pKa = 11.84HH483 pKa = 5.66TLDD486 pKa = 5.29RR487 pKa = 11.84ILPRR491 pKa = 11.84IFGPSTDD498 pKa = 3.46TVFDD502 pKa = 4.96FIEE505 pKa = 4.19SAITSSWVSIDD516 pKa = 2.85ARR518 pKa = 11.84KK519 pKa = 10.06RR520 pKa = 11.84NGRR523 pKa = 11.84ARR525 pKa = 11.84KK526 pKa = 9.01FRR528 pKa = 11.84TAFINRR534 pKa = 11.84FHH536 pKa = 7.21DD537 pKa = 3.54PEE539 pKa = 3.96FAYY542 pKa = 10.14MFGITGNGIEE552 pKa = 4.07RR553 pKa = 11.84MEE555 pKa = 4.37GKK557 pKa = 8.6VTSNIAQEE565 pKa = 3.93VDD567 pKa = 3.16YY568 pKa = 11.37LMNGGDD574 pKa = 4.79LRR576 pKa = 11.84NCPILRR582 pKa = 11.84TLKK585 pKa = 9.83AAEE588 pKa = 4.14RR589 pKa = 11.84EE590 pKa = 4.18EE591 pKa = 4.72TVTFMCKK598 pKa = 9.82EE599 pKa = 4.18KK600 pKa = 10.96VGSLYY605 pKa = 10.78AIDD608 pKa = 3.41GTVRR612 pKa = 11.84VFKK615 pKa = 10.72RR616 pKa = 11.84FEE618 pKa = 4.46TIDD621 pKa = 3.6LAQLGWTSHH630 pKa = 4.96GKK632 pKa = 9.31VMKK635 pKa = 9.69PYY637 pKa = 10.74AFRR640 pKa = 11.84APVIQGMTICSTAYY654 pKa = 8.86TSTAIDD660 pKa = 3.78IITTVFGPLRR670 pKa = 11.84LRR672 pKa = 11.84VGSLFEE678 pKa = 3.97
MM1 pKa = 7.19EE2 pKa = 5.64ASANGLSHH10 pKa = 7.93DD11 pKa = 4.42DD12 pKa = 4.09NANKK16 pKa = 9.65SQNVGPSTLPRR27 pKa = 11.84SDD29 pKa = 3.14KK30 pKa = 11.09QGGEE34 pKa = 3.64KK35 pKa = 10.34HH36 pKa = 6.28EE37 pKa = 4.3NSFNSFSNDD46 pKa = 2.5FFFNFLRR53 pKa = 11.84MSTNTHH59 pKa = 6.17ISDD62 pKa = 3.74SPGVSFVAKK71 pKa = 10.42DD72 pKa = 3.41GTPYY76 pKa = 10.95SSLTIPSGVGRR87 pKa = 11.84LTHH90 pKa = 6.16NVVASAVQLNITASNTLEE108 pKa = 3.65VDD110 pKa = 3.38YY111 pKa = 11.5GFGQDD116 pKa = 3.26VSRR119 pKa = 11.84TTGTIPIPIFDD130 pKa = 3.77GEE132 pKa = 4.61KK133 pKa = 9.59YY134 pKa = 10.29KK135 pKa = 10.09EE136 pKa = 4.0TARR139 pKa = 11.84ALAAIFSKK147 pKa = 10.62KK148 pKa = 10.06GMSVDD153 pKa = 3.52VTSQTVQEE161 pKa = 4.23TLKK164 pKa = 11.21NSDD167 pKa = 3.33LTIATVAAGYY177 pKa = 7.73YY178 pKa = 7.0TALAARR184 pKa = 11.84HH185 pKa = 5.96EE186 pKa = 4.28LTKK189 pKa = 10.79DD190 pKa = 3.28VSEE193 pKa = 4.52AAHH196 pKa = 6.69TIPFVTALSDD206 pKa = 3.54TFSAALNAQRR216 pKa = 11.84TSHH219 pKa = 6.43VISSCLRR226 pKa = 11.84CPNSRR231 pKa = 11.84NAQRR235 pKa = 11.84DD236 pKa = 3.62IVIGTVLWNNVFVEE250 pKa = 4.4SLSEE254 pKa = 3.98HH255 pKa = 5.97NMAVPNPNDD264 pKa = 2.91ISFFIPNKK272 pKa = 9.99ALSSSWWCAIWLLNAFLHH290 pKa = 6.5SFIAPTRR297 pKa = 11.84IHH299 pKa = 6.94IFITQGEE306 pKa = 4.94TYY308 pKa = 10.07HH309 pKa = 6.92LAPFTDD315 pKa = 3.16SDD317 pKa = 3.84VYY319 pKa = 10.42EE320 pKa = 4.07AVRR323 pKa = 11.84FLLAMSKK330 pKa = 10.16SSRR333 pKa = 11.84PMPEE337 pKa = 3.86SVEE340 pKa = 4.31SMLYY344 pKa = 10.39AYY346 pKa = 7.32GTQMIIQPHH355 pKa = 4.82SLYY358 pKa = 10.63TEE360 pKa = 4.42GGLIRR365 pKa = 11.84RR366 pKa = 11.84MIFTVPHH373 pKa = 6.59LPAHH377 pKa = 6.9GYY379 pKa = 9.65FVTNSEE385 pKa = 4.1FSRR388 pKa = 11.84YY389 pKa = 7.39MNIAVPDD396 pKa = 4.1DD397 pKa = 3.67PRR399 pKa = 11.84SAKK402 pKa = 10.69DD403 pKa = 3.21FVIGAGTGLLQIVLAYY419 pKa = 9.72QAAFSCAGPIALHH432 pKa = 5.63WHH434 pKa = 6.29ANDD437 pKa = 5.58AISQGMDD444 pKa = 2.8RR445 pKa = 11.84VASIYY450 pKa = 10.99LEE452 pKa = 3.79GRR454 pKa = 11.84YY455 pKa = 7.58FTIPMAVNVATNVAQYY471 pKa = 7.91TTMVRR476 pKa = 11.84ADD478 pKa = 3.19PEE480 pKa = 4.18YY481 pKa = 10.68RR482 pKa = 11.84HH483 pKa = 5.66TLDD486 pKa = 5.29RR487 pKa = 11.84ILPRR491 pKa = 11.84IFGPSTDD498 pKa = 3.46TVFDD502 pKa = 4.96FIEE505 pKa = 4.19SAITSSWVSIDD516 pKa = 2.85ARR518 pKa = 11.84KK519 pKa = 10.06RR520 pKa = 11.84NGRR523 pKa = 11.84ARR525 pKa = 11.84KK526 pKa = 9.01FRR528 pKa = 11.84TAFINRR534 pKa = 11.84FHH536 pKa = 7.21DD537 pKa = 3.54PEE539 pKa = 3.96FAYY542 pKa = 10.14MFGITGNGIEE552 pKa = 4.07RR553 pKa = 11.84MEE555 pKa = 4.37GKK557 pKa = 8.6VTSNIAQEE565 pKa = 3.93VDD567 pKa = 3.16YY568 pKa = 11.37LMNGGDD574 pKa = 4.79LRR576 pKa = 11.84NCPILRR582 pKa = 11.84TLKK585 pKa = 9.83AAEE588 pKa = 4.14RR589 pKa = 11.84EE590 pKa = 4.18EE591 pKa = 4.72TVTFMCKK598 pKa = 9.82EE599 pKa = 4.18KK600 pKa = 10.96VGSLYY605 pKa = 10.78AIDD608 pKa = 3.41GTVRR612 pKa = 11.84VFKK615 pKa = 10.72RR616 pKa = 11.84FEE618 pKa = 4.46TIDD621 pKa = 3.6LAQLGWTSHH630 pKa = 4.96GKK632 pKa = 9.31VMKK635 pKa = 9.69PYY637 pKa = 10.74AFRR640 pKa = 11.84APVIQGMTICSTAYY654 pKa = 8.86TSTAIDD660 pKa = 3.78IITTVFGPLRR670 pKa = 11.84LRR672 pKa = 11.84VGSLFEE678 pKa = 3.97
Molecular weight: 74.92 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q90155|Q90155_TTV1 RNA-directed RNA polymerase (Fragment) OS=Trichomonas vaginalis virus 1 OX=674953 GN=pol PE=3 SV=1
DDD2 pKa = 4.78FLSKKK7 pKa = 10.49VRR9 pKa = 11.84CGPVIPSVKKK19 pKa = 9.74HH20 pKa = 5.5HH21 pKa = 6.27NINYYY26 pKa = 10.17SILKKK31 pKa = 10.49HH32 pKa = 6.04GNEEE36 pKa = 3.99YY37 pKa = 9.7FVPGYYY43 pKa = 10.73WVLQDDD49 pKa = 5.65DD50 pKa = 4.48YY51 pKa = 11.87LNAVKKK57 pKa = 9.93VGEEE61 pKa = 4.51DDD63 pKa = 3.98PPNQLPYYY71 pKa = 11.11DD72 pKa = 4.63DD73 pKa = 6.11DD74 pKa = 5.37LFTYYY79 pKa = 10.56KKK81 pKa = 9.93LLYYY85 pKa = 10.41DD86 pKa = 4.23YY87 pKa = 10.51SHHH90 pKa = 6.81PEEE93 pKa = 4.23RR94 pKa = 11.84HHH96 pKa = 5.87KK97 pKa = 10.72PRR99 pKa = 11.84LLTSEEE105 pKa = 4.73EEE107 pKa = 4.08QLFPLKKK114 pKa = 10.3EE115 pKa = 3.76SAARR119 pKa = 11.84TKKK122 pKa = 11.23NFYYY126 pKa = 11.3RR127 pKa = 11.84TLWNEEE133 pKa = 3.91TSDDD137 pKa = 3.13KK138 pKa = 11.34AFKKK142 pKa = 10.5GTYYY146 pKa = 10.11DDD148 pKa = 3.91VAGLLMWQQCALMWSLPKKK167 pKa = 10.65IINKKK172 pKa = 9.51ISGVCDDD179 pKa = 3.43LTEEE183 pKa = 4.38KK184 pKa = 10.97SLTLLKKK191 pKa = 10.49ISDDD195 pKa = 3.64LKKK198 pKa = 10.38LGLAYYY204 pKa = 10.64PIFRR208 pKa = 11.84LFIEEE213 pKa = 4.84PTLLGRR219 pKa = 11.84GAIPGDDD226 pKa = 3.15ALDDD230 pKa = 3.59KKK232 pKa = 10.91HH233 pKa = 6.52LTYYY237 pKa = 10.29PLMTVDDD244 pKa = 3.57PKKK247 pKa = 9.7QLHHH251 pKa = 5.85DD252 pKa = 4.82IYYY255 pKa = 10.84LLSRR259 pKa = 11.84NYYY262 pKa = 9.41NTKKK266 pKa = 9.54SSFEEE271 pKa = 3.66HH272 pKa = 6.5HH273 pKa = 6.74EEE275 pKa = 4.18EE276 pKa = 4.43LLWSRR281 pKa = 11.84SGSHHH286 pKa = 6.32YY287 pKa = 11.13YY288 pKa = 9.85DDD290 pKa = 3.61EE291 pKa = 4.81IDDD294 pKa = 3.8LLPPQPTRR302 pKa = 11.84KKK304 pKa = 9.37EE305 pKa = 3.75LDDD308 pKa = 3.72VTIDDD313 pKa = 4.02YY314 pKa = 10.63KKK316 pKa = 9.99CKKK319 pKa = 7.74QVFIRR324 pKa = 11.84QSRR327 pKa = 11.84KKK329 pKa = 9.69EEE331 pKa = 4.08HH332 pKa = 5.88KKK334 pKa = 9.68EE335 pKa = 3.72FIYYY339 pKa = 10.13CDDD342 pKa = 3.73ISYYY346 pKa = 9.69YYY348 pKa = 10.58DDD350 pKa = 4.24VLKKK354 pKa = 10.68FEEE357 pKa = 5.38GWQDDD362 pKa = 2.84EEE364 pKa = 5.9ILSPGDDD371 pKa = 3.6YY372 pKa = 10.61SEEE375 pKa = 3.95LHHH378 pKa = 6.73KKK380 pKa = 9.77SGYYY384 pKa = 9.98KK385 pKa = 8.42YY386 pKa = 10.71KK387 pKa = 11.29MLDDD391 pKa = 3.61YY392 pKa = 11.3DDD394 pKa = 5.84NSQHHH399 pKa = 6.71IQSMRR404 pKa = 11.84LIFEEE409 pKa = 4.35MKKK412 pKa = 10.79EE413 pKa = 4.04LPPEEE418 pKa = 4.42SFALDDD424 pKa = 3.01CIASFDDD431 pKa = 3.88MQTSDDD437 pKa = 3.33RR438 pKa = 11.84KKK440 pKa = 5.64TATLPSGHHH449 pKa = 7.21ATTFINTVLNWCYYY463 pKa = 9.23QMVGLKKK470 pKa = 9.52DDD472 pKa = 4.16FMCAGDDD479 pKa = 4.46DD480 pKa = 4.1ILMSQEEE487 pKa = 4.57ISLAPILKKK496 pKa = 10.02QFKKK500 pKa = 10.32NPSKKK505 pKa = 10.84STGTRR510 pKa = 11.84GEEE513 pKa = 4.34LRR515 pKa = 11.84KKK517 pKa = 9.45HH518 pKa = 4.23YY519 pKa = 10.01EEE521 pKa = 5.28GVFAYYY527 pKa = 8.75CRR529 pKa = 11.84AIASLVSGNWLSEEE543 pKa = 4.12LRR545 pKa = 11.84DDD547 pKa = 3.6TPILVPIQNGIDDD560 pKa = 3.56LRR562 pKa = 11.84SRR564 pKa = 11.84AGLLGVPWKKK574 pKa = 10.76GLSEEE579 pKa = 4.86IEEE582 pKa = 4.55EEE584 pKa = 4.51IPKKK588 pKa = 9.28DD589 pKa = 3.53SMALLNSHHH598 pKa = 6.99AGPGLITRR606 pKa = 11.84DDD608 pKa = 3.58YY609 pKa = 11.53SFTVTPTPPKKK620 pKa = 9.61HHH622 pKa = 6.77SLEEE626 pKa = 4.23YY627 pKa = 9.01ATRR630 pKa = 11.84HHH632 pKa = 6.01LQDDD636 pKa = 3.62CKKK639 pKa = 9.78HH640 pKa = 5.67PWKKK644 pKa = 10.56LTANEEE650 pKa = 4.37NKKK653 pKa = 10.3GQQIKKK659 pKa = 10.35KK660 pKa = 8.89SHHH663 pKa = 5.28HHH665 pKa = 5.12SQTKKK670 pKa = 7.74TYYY673 pKa = 8.04KK674 pKa = 10.31VYYY677 pKa = 9.85EE678 pKa = 4.11FKKK681 pKa = 10.82SGLPTVLSEEE691 pKa = 4.36SQSALSLVWWQAMLKKK707 pKa = 10.25EE708 pKa = 4.09MQDDD712 pKa = 3.36YY713 pKa = 8.34TKKK716 pKa = 10.71KK717 pKa = 10.21KK718 pKa = 9.84DD719 pKa = 3.19HHH721 pKa = 6.25YYY723 pKa = 9.96CNACTSSVSGDDD735 pKa = 3.03FLRR738 pKa = 11.84ATSKKK743 pKa = 10.11AGVLITSLISSSS
DDD2 pKa = 4.78FLSKKK7 pKa = 10.49VRR9 pKa = 11.84CGPVIPSVKKK19 pKa = 9.74HH20 pKa = 5.5HH21 pKa = 6.27NINYYY26 pKa = 10.17SILKKK31 pKa = 10.49HH32 pKa = 6.04GNEEE36 pKa = 3.99YY37 pKa = 9.7FVPGYYY43 pKa = 10.73WVLQDDD49 pKa = 5.65DD50 pKa = 4.48YY51 pKa = 11.87LNAVKKK57 pKa = 9.93VGEEE61 pKa = 4.51DDD63 pKa = 3.98PPNQLPYYY71 pKa = 11.11DD72 pKa = 4.63DD73 pKa = 6.11DD74 pKa = 5.37LFTYYY79 pKa = 10.56KKK81 pKa = 9.93LLYYY85 pKa = 10.41DD86 pKa = 4.23YY87 pKa = 10.51SHHH90 pKa = 6.81PEEE93 pKa = 4.23RR94 pKa = 11.84HHH96 pKa = 5.87KK97 pKa = 10.72PRR99 pKa = 11.84LLTSEEE105 pKa = 4.73EEE107 pKa = 4.08QLFPLKKK114 pKa = 10.3EE115 pKa = 3.76SAARR119 pKa = 11.84TKKK122 pKa = 11.23NFYYY126 pKa = 11.3RR127 pKa = 11.84TLWNEEE133 pKa = 3.91TSDDD137 pKa = 3.13KK138 pKa = 11.34AFKKK142 pKa = 10.5GTYYY146 pKa = 10.11DDD148 pKa = 3.91VAGLLMWQQCALMWSLPKKK167 pKa = 10.65IINKKK172 pKa = 9.51ISGVCDDD179 pKa = 3.43LTEEE183 pKa = 4.38KK184 pKa = 10.97SLTLLKKK191 pKa = 10.49ISDDD195 pKa = 3.64LKKK198 pKa = 10.38LGLAYYY204 pKa = 10.64PIFRR208 pKa = 11.84LFIEEE213 pKa = 4.84PTLLGRR219 pKa = 11.84GAIPGDDD226 pKa = 3.15ALDDD230 pKa = 3.59KKK232 pKa = 10.91HH233 pKa = 6.52LTYYY237 pKa = 10.29PLMTVDDD244 pKa = 3.57PKKK247 pKa = 9.7QLHHH251 pKa = 5.85DD252 pKa = 4.82IYYY255 pKa = 10.84LLSRR259 pKa = 11.84NYYY262 pKa = 9.41NTKKK266 pKa = 9.54SSFEEE271 pKa = 3.66HH272 pKa = 6.5HH273 pKa = 6.74EEE275 pKa = 4.18EE276 pKa = 4.43LLWSRR281 pKa = 11.84SGSHHH286 pKa = 6.32YY287 pKa = 11.13YY288 pKa = 9.85DDD290 pKa = 3.61EE291 pKa = 4.81IDDD294 pKa = 3.8LLPPQPTRR302 pKa = 11.84KKK304 pKa = 9.37EE305 pKa = 3.75LDDD308 pKa = 3.72VTIDDD313 pKa = 4.02YY314 pKa = 10.63KKK316 pKa = 9.99CKKK319 pKa = 7.74QVFIRR324 pKa = 11.84QSRR327 pKa = 11.84KKK329 pKa = 9.69EEE331 pKa = 4.08HH332 pKa = 5.88KKK334 pKa = 9.68EE335 pKa = 3.72FIYYY339 pKa = 10.13CDDD342 pKa = 3.73ISYYY346 pKa = 9.69YYY348 pKa = 10.58DDD350 pKa = 4.24VLKKK354 pKa = 10.68FEEE357 pKa = 5.38GWQDDD362 pKa = 2.84EEE364 pKa = 5.9ILSPGDDD371 pKa = 3.6YY372 pKa = 10.61SEEE375 pKa = 3.95LHHH378 pKa = 6.73KKK380 pKa = 9.77SGYYY384 pKa = 9.98KK385 pKa = 8.42YY386 pKa = 10.71KK387 pKa = 11.29MLDDD391 pKa = 3.61YY392 pKa = 11.3DDD394 pKa = 5.84NSQHHH399 pKa = 6.71IQSMRR404 pKa = 11.84LIFEEE409 pKa = 4.35MKKK412 pKa = 10.79EE413 pKa = 4.04LPPEEE418 pKa = 4.42SFALDDD424 pKa = 3.01CIASFDDD431 pKa = 3.88MQTSDDD437 pKa = 3.33RR438 pKa = 11.84KKK440 pKa = 5.64TATLPSGHHH449 pKa = 7.21ATTFINTVLNWCYYY463 pKa = 9.23QMVGLKKK470 pKa = 9.52DDD472 pKa = 4.16FMCAGDDD479 pKa = 4.46DD480 pKa = 4.1ILMSQEEE487 pKa = 4.57ISLAPILKKK496 pKa = 10.02QFKKK500 pKa = 10.32NPSKKK505 pKa = 10.84STGTRR510 pKa = 11.84GEEE513 pKa = 4.34LRR515 pKa = 11.84KKK517 pKa = 9.45HH518 pKa = 4.23YY519 pKa = 10.01EEE521 pKa = 5.28GVFAYYY527 pKa = 8.75CRR529 pKa = 11.84AIASLVSGNWLSEEE543 pKa = 4.12LRR545 pKa = 11.84DDD547 pKa = 3.6TPILVPIQNGIDDD560 pKa = 3.56LRR562 pKa = 11.84SRR564 pKa = 11.84AGLLGVPWKKK574 pKa = 10.76GLSEEE579 pKa = 4.86IEEE582 pKa = 4.55EEE584 pKa = 4.51IPKKK588 pKa = 9.28DD589 pKa = 3.53SMALLNSHHH598 pKa = 6.99AGPGLITRR606 pKa = 11.84DDD608 pKa = 3.58YY609 pKa = 11.53SFTVTPTPPKKK620 pKa = 9.61HHH622 pKa = 6.77SLEEE626 pKa = 4.23YY627 pKa = 9.01ATRR630 pKa = 11.84HHH632 pKa = 6.01LQDDD636 pKa = 3.62CKKK639 pKa = 9.78HH640 pKa = 5.67PWKKK644 pKa = 10.56LTANEEE650 pKa = 4.37NKKK653 pKa = 10.3GQQIKKK659 pKa = 10.35KK660 pKa = 8.89SHHH663 pKa = 5.28HHH665 pKa = 5.12SQTKKK670 pKa = 7.74TYYY673 pKa = 8.04KK674 pKa = 10.31VYYY677 pKa = 9.85EE678 pKa = 4.11FKKK681 pKa = 10.82SGLPTVLSEEE691 pKa = 4.36SQSALSLVWWQAMLKKK707 pKa = 10.25EE708 pKa = 4.09MQDDD712 pKa = 3.36YY713 pKa = 8.34TKKK716 pKa = 10.71KK717 pKa = 10.21KK718 pKa = 9.84DD719 pKa = 3.19HHH721 pKa = 6.25YYY723 pKa = 9.96CNACTSSVSGDDD735 pKa = 3.03FLRR738 pKa = 11.84ATSKKK743 pKa = 10.11AGVLITSLISSSS
Molecular weight: 85.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1434 |
678 |
756 |
717.0 |
80.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.531 ± 1.119 | 1.604 ± 0.363 |
5.091 ± 0.142 | 4.463 ± 0.024 |
5.021 ± 0.559 | 5.439 ± 0.386 |
2.859 ± 0.036 | 6.206 ± 0.461 |
5.021 ± 1.129 | 9.484 ± 1.81 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.58 ± 0.141 | 4.393 ± 0.395 |
5.021 ± 0.285 | 3.347 ± 0.44 |
4.881 ± 0.46 | 8.926 ± 0.049 |
7.252 ± 0.64 | 5.509 ± 0.811 |
1.534 ± 0.319 | 3.835 ± 0.375 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
