
Hubei narna-like virus 19
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.87
Get precalculated fractions of proteins
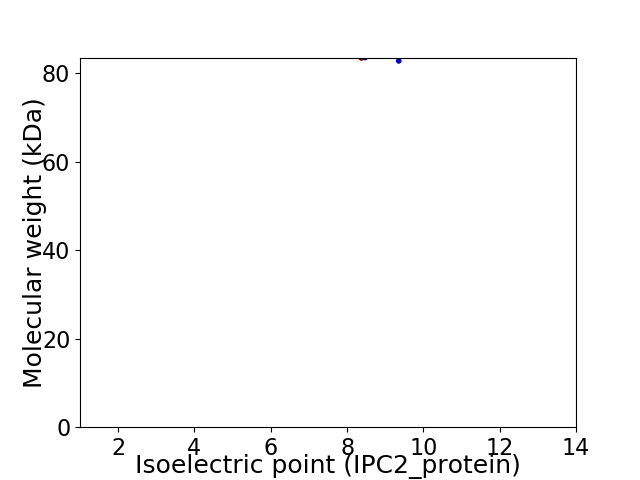
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
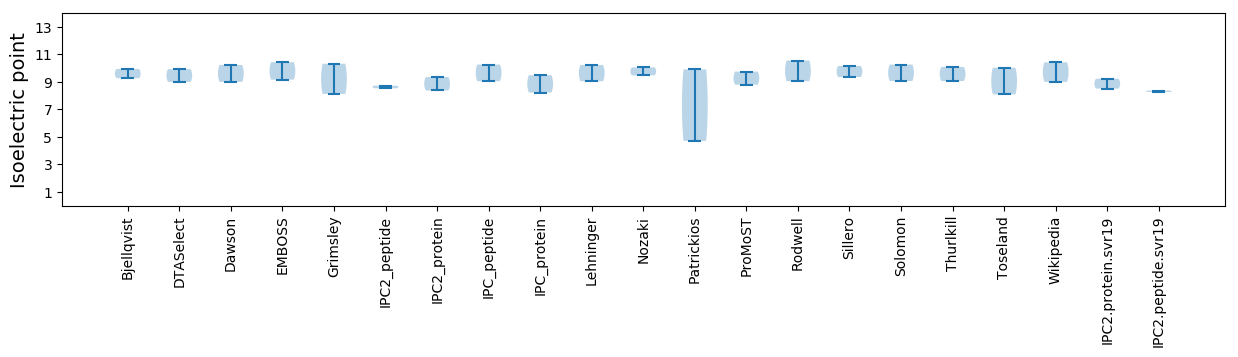
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KIH1|A0A1L3KIH1_9VIRU RNA-dependent RNA polymerase OS=Hubei narna-like virus 19 OX=1922949 PE=4 SV=1
MM1 pKa = 7.24SGAMPKK7 pKa = 9.5HH8 pKa = 5.88CEE10 pKa = 3.85VRR12 pKa = 11.84KK13 pKa = 8.53VTLAALCRR21 pKa = 11.84PLVCTSSGRR30 pKa = 11.84LGVWDD35 pKa = 3.67VRR37 pKa = 11.84SGHH40 pKa = 6.56RR41 pKa = 11.84SDD43 pKa = 4.13PVRR46 pKa = 11.84KK47 pKa = 9.34SGRR50 pKa = 11.84VRR52 pKa = 11.84IRR54 pKa = 11.84SWDD57 pKa = 3.59PPRR60 pKa = 11.84LYY62 pKa = 10.01PGQCHH67 pKa = 5.81EE68 pKa = 4.6PRR70 pKa = 11.84APLAEE75 pKa = 4.61ADD77 pKa = 3.61GATRR81 pKa = 11.84TVSMSEE87 pKa = 3.88VTPITASSLIEE98 pKa = 4.14LGLLRR103 pKa = 11.84FGGPDD108 pKa = 3.14GLGPLLAEE116 pKa = 3.92VFLRR120 pKa = 11.84EE121 pKa = 4.44NIHH124 pKa = 5.75EE125 pKa = 4.43TTGDD129 pKa = 3.4AVRR132 pKa = 11.84SCDD135 pKa = 3.54EE136 pKa = 3.99TCQLDD141 pKa = 3.87SCLMMTGRR149 pKa = 11.84VTYY152 pKa = 10.28SGPNRR157 pKa = 11.84EE158 pKa = 4.54RR159 pKa = 11.84YY160 pKa = 7.8TIQPHH165 pKa = 5.91PAFSIWQTGSTQIQRR180 pKa = 11.84EE181 pKa = 4.3MWWNSSGEE189 pKa = 4.16PNHH192 pKa = 6.53KK193 pKa = 9.08VTVIQPYY200 pKa = 9.87GSVRR204 pKa = 11.84LAALARR210 pKa = 11.84YY211 pKa = 8.06YY212 pKa = 10.01IRR214 pKa = 11.84SLGEE218 pKa = 3.68PSSQGLNQGALTGSRR233 pKa = 11.84TNYY236 pKa = 9.72ASNRR240 pKa = 11.84KK241 pKa = 8.1VLKK244 pKa = 10.06DD245 pKa = 3.31LHH247 pKa = 7.13AGYY250 pKa = 10.24YY251 pKa = 9.87INWDD255 pKa = 3.65EE256 pKa = 4.22LTQGSLQGVGKK267 pKa = 9.21LLCSEE272 pKa = 4.73TVRR275 pKa = 11.84EE276 pKa = 4.16THH278 pKa = 6.76CPLGKK283 pKa = 9.91SAAGARR289 pKa = 11.84NDD291 pKa = 3.23SFRR294 pKa = 11.84PVYY297 pKa = 10.79GEE299 pKa = 3.56AHH301 pKa = 7.08LLHH304 pKa = 7.2DD305 pKa = 3.78IEE307 pKa = 5.16VNFRR311 pKa = 11.84PWEE314 pKa = 4.35DD315 pKa = 3.63PEE317 pKa = 4.5EE318 pKa = 4.41VIPSDD323 pKa = 3.18NMRR326 pKa = 11.84VRR328 pKa = 11.84TISSFLHH335 pKa = 5.78AKK337 pKa = 9.95SVEE340 pKa = 4.03KK341 pKa = 10.96GKK343 pKa = 11.18GEE345 pKa = 3.79EE346 pKa = 4.36AEE348 pKa = 4.73RR349 pKa = 11.84SAHH352 pKa = 5.94MNTAGTRR359 pKa = 11.84DD360 pKa = 3.8DD361 pKa = 3.45GTIRR365 pKa = 11.84ITEE368 pKa = 3.94LHH370 pKa = 6.16RR371 pKa = 11.84AKK373 pKa = 10.33HH374 pKa = 5.92DD375 pKa = 3.49SAVRR379 pKa = 11.84INVRR383 pKa = 11.84HH384 pKa = 4.91NMMWLEE390 pKa = 3.8EE391 pKa = 4.29SGPEE395 pKa = 3.58RR396 pKa = 11.84LRR398 pKa = 11.84INVGNVVCCIGKK410 pKa = 8.41VTANADD416 pKa = 3.71TVPVEE421 pKa = 4.13RR422 pKa = 11.84RR423 pKa = 11.84PNGAKK428 pKa = 9.91KK429 pKa = 10.69LNIIGLSIHH438 pKa = 6.15VGPRR442 pKa = 11.84LIRR445 pKa = 11.84HH446 pKa = 6.22HH447 pKa = 6.81CPHH450 pKa = 6.99LSSYY454 pKa = 11.1YY455 pKa = 8.42ITNDD459 pKa = 3.07EE460 pKa = 4.24GSSGEE465 pKa = 3.96LCKK468 pKa = 10.73NPRR471 pKa = 11.84FISFFRR477 pKa = 11.84DD478 pKa = 2.89RR479 pKa = 11.84HH480 pKa = 5.43HH481 pKa = 7.16VGNPLLLWRR490 pKa = 11.84DD491 pKa = 3.54GTSCHH496 pKa = 6.69IDD498 pKa = 3.31SVVKK502 pKa = 10.13GFRR505 pKa = 11.84DD506 pKa = 3.56PRR508 pKa = 11.84VEE510 pKa = 3.27IALRR514 pKa = 11.84KK515 pKa = 9.41RR516 pKa = 11.84FHH518 pKa = 6.11TFEE521 pKa = 3.73IALRR525 pKa = 11.84FCIADD530 pKa = 3.63QLLHH534 pKa = 6.15TSSFEE539 pKa = 4.16DD540 pKa = 3.15RR541 pKa = 11.84CSLRR545 pKa = 11.84RR546 pKa = 11.84GVLIRR551 pKa = 11.84CPRR554 pKa = 11.84NFLFKK559 pKa = 10.42EE560 pKa = 3.8VGIISRR566 pKa = 11.84EE567 pKa = 3.91LPGPIVNVVGQFHH580 pKa = 7.2VDD582 pKa = 3.27ITWRR586 pKa = 11.84SCDD589 pKa = 3.55DD590 pKa = 3.97RR591 pKa = 11.84SVCLYY596 pKa = 11.17GLGLDD601 pKa = 3.23KK602 pKa = 10.92CIPFRR607 pKa = 11.84EE608 pKa = 4.33TSANARR614 pKa = 11.84NLRR617 pKa = 11.84PSLGVAHH624 pKa = 6.77VPGQPFLEE632 pKa = 4.87GTRR635 pKa = 11.84HH636 pKa = 3.64VTLRR640 pKa = 11.84SGSSTSHH647 pKa = 6.53RR648 pKa = 11.84PLAVLEE654 pKa = 4.42AEE656 pKa = 4.41LLQLLGYY663 pKa = 8.3VVPTPRR669 pKa = 11.84VTRR672 pKa = 11.84TAEE675 pKa = 4.0EE676 pKa = 4.27PTCEE680 pKa = 4.0YY681 pKa = 10.74LCRR684 pKa = 11.84DD685 pKa = 5.08DD686 pKa = 4.45ISKK689 pKa = 10.78LLVVSKK695 pKa = 10.81YY696 pKa = 11.02NIGHH700 pKa = 6.41NSFPDD705 pKa = 3.14RR706 pKa = 11.84VRR708 pKa = 11.84ISNSIPIRR716 pKa = 11.84QGRR719 pKa = 11.84HH720 pKa = 3.53WQLMPMSCPNNTRR733 pKa = 11.84GNRR736 pKa = 11.84PRR738 pKa = 11.84QYY740 pKa = 11.05VRR742 pKa = 11.84WW743 pKa = 3.79
MM1 pKa = 7.24SGAMPKK7 pKa = 9.5HH8 pKa = 5.88CEE10 pKa = 3.85VRR12 pKa = 11.84KK13 pKa = 8.53VTLAALCRR21 pKa = 11.84PLVCTSSGRR30 pKa = 11.84LGVWDD35 pKa = 3.67VRR37 pKa = 11.84SGHH40 pKa = 6.56RR41 pKa = 11.84SDD43 pKa = 4.13PVRR46 pKa = 11.84KK47 pKa = 9.34SGRR50 pKa = 11.84VRR52 pKa = 11.84IRR54 pKa = 11.84SWDD57 pKa = 3.59PPRR60 pKa = 11.84LYY62 pKa = 10.01PGQCHH67 pKa = 5.81EE68 pKa = 4.6PRR70 pKa = 11.84APLAEE75 pKa = 4.61ADD77 pKa = 3.61GATRR81 pKa = 11.84TVSMSEE87 pKa = 3.88VTPITASSLIEE98 pKa = 4.14LGLLRR103 pKa = 11.84FGGPDD108 pKa = 3.14GLGPLLAEE116 pKa = 3.92VFLRR120 pKa = 11.84EE121 pKa = 4.44NIHH124 pKa = 5.75EE125 pKa = 4.43TTGDD129 pKa = 3.4AVRR132 pKa = 11.84SCDD135 pKa = 3.54EE136 pKa = 3.99TCQLDD141 pKa = 3.87SCLMMTGRR149 pKa = 11.84VTYY152 pKa = 10.28SGPNRR157 pKa = 11.84EE158 pKa = 4.54RR159 pKa = 11.84YY160 pKa = 7.8TIQPHH165 pKa = 5.91PAFSIWQTGSTQIQRR180 pKa = 11.84EE181 pKa = 4.3MWWNSSGEE189 pKa = 4.16PNHH192 pKa = 6.53KK193 pKa = 9.08VTVIQPYY200 pKa = 9.87GSVRR204 pKa = 11.84LAALARR210 pKa = 11.84YY211 pKa = 8.06YY212 pKa = 10.01IRR214 pKa = 11.84SLGEE218 pKa = 3.68PSSQGLNQGALTGSRR233 pKa = 11.84TNYY236 pKa = 9.72ASNRR240 pKa = 11.84KK241 pKa = 8.1VLKK244 pKa = 10.06DD245 pKa = 3.31LHH247 pKa = 7.13AGYY250 pKa = 10.24YY251 pKa = 9.87INWDD255 pKa = 3.65EE256 pKa = 4.22LTQGSLQGVGKK267 pKa = 9.21LLCSEE272 pKa = 4.73TVRR275 pKa = 11.84EE276 pKa = 4.16THH278 pKa = 6.76CPLGKK283 pKa = 9.91SAAGARR289 pKa = 11.84NDD291 pKa = 3.23SFRR294 pKa = 11.84PVYY297 pKa = 10.79GEE299 pKa = 3.56AHH301 pKa = 7.08LLHH304 pKa = 7.2DD305 pKa = 3.78IEE307 pKa = 5.16VNFRR311 pKa = 11.84PWEE314 pKa = 4.35DD315 pKa = 3.63PEE317 pKa = 4.5EE318 pKa = 4.41VIPSDD323 pKa = 3.18NMRR326 pKa = 11.84VRR328 pKa = 11.84TISSFLHH335 pKa = 5.78AKK337 pKa = 9.95SVEE340 pKa = 4.03KK341 pKa = 10.96GKK343 pKa = 11.18GEE345 pKa = 3.79EE346 pKa = 4.36AEE348 pKa = 4.73RR349 pKa = 11.84SAHH352 pKa = 5.94MNTAGTRR359 pKa = 11.84DD360 pKa = 3.8DD361 pKa = 3.45GTIRR365 pKa = 11.84ITEE368 pKa = 3.94LHH370 pKa = 6.16RR371 pKa = 11.84AKK373 pKa = 10.33HH374 pKa = 5.92DD375 pKa = 3.49SAVRR379 pKa = 11.84INVRR383 pKa = 11.84HH384 pKa = 4.91NMMWLEE390 pKa = 3.8EE391 pKa = 4.29SGPEE395 pKa = 3.58RR396 pKa = 11.84LRR398 pKa = 11.84INVGNVVCCIGKK410 pKa = 8.41VTANADD416 pKa = 3.71TVPVEE421 pKa = 4.13RR422 pKa = 11.84RR423 pKa = 11.84PNGAKK428 pKa = 9.91KK429 pKa = 10.69LNIIGLSIHH438 pKa = 6.15VGPRR442 pKa = 11.84LIRR445 pKa = 11.84HH446 pKa = 6.22HH447 pKa = 6.81CPHH450 pKa = 6.99LSSYY454 pKa = 11.1YY455 pKa = 8.42ITNDD459 pKa = 3.07EE460 pKa = 4.24GSSGEE465 pKa = 3.96LCKK468 pKa = 10.73NPRR471 pKa = 11.84FISFFRR477 pKa = 11.84DD478 pKa = 2.89RR479 pKa = 11.84HH480 pKa = 5.43HH481 pKa = 7.16VGNPLLLWRR490 pKa = 11.84DD491 pKa = 3.54GTSCHH496 pKa = 6.69IDD498 pKa = 3.31SVVKK502 pKa = 10.13GFRR505 pKa = 11.84DD506 pKa = 3.56PRR508 pKa = 11.84VEE510 pKa = 3.27IALRR514 pKa = 11.84KK515 pKa = 9.41RR516 pKa = 11.84FHH518 pKa = 6.11TFEE521 pKa = 3.73IALRR525 pKa = 11.84FCIADD530 pKa = 3.63QLLHH534 pKa = 6.15TSSFEE539 pKa = 4.16DD540 pKa = 3.15RR541 pKa = 11.84CSLRR545 pKa = 11.84RR546 pKa = 11.84GVLIRR551 pKa = 11.84CPRR554 pKa = 11.84NFLFKK559 pKa = 10.42EE560 pKa = 3.8VGIISRR566 pKa = 11.84EE567 pKa = 3.91LPGPIVNVVGQFHH580 pKa = 7.2VDD582 pKa = 3.27ITWRR586 pKa = 11.84SCDD589 pKa = 3.55DD590 pKa = 3.97RR591 pKa = 11.84SVCLYY596 pKa = 11.17GLGLDD601 pKa = 3.23KK602 pKa = 10.92CIPFRR607 pKa = 11.84EE608 pKa = 4.33TSANARR614 pKa = 11.84NLRR617 pKa = 11.84PSLGVAHH624 pKa = 6.77VPGQPFLEE632 pKa = 4.87GTRR635 pKa = 11.84HH636 pKa = 3.64VTLRR640 pKa = 11.84SGSSTSHH647 pKa = 6.53RR648 pKa = 11.84PLAVLEE654 pKa = 4.42AEE656 pKa = 4.41LLQLLGYY663 pKa = 8.3VVPTPRR669 pKa = 11.84VTRR672 pKa = 11.84TAEE675 pKa = 4.0EE676 pKa = 4.27PTCEE680 pKa = 4.0YY681 pKa = 10.74LCRR684 pKa = 11.84DD685 pKa = 5.08DD686 pKa = 4.45ISKK689 pKa = 10.78LLVVSKK695 pKa = 10.81YY696 pKa = 11.02NIGHH700 pKa = 6.41NSFPDD705 pKa = 3.14RR706 pKa = 11.84VRR708 pKa = 11.84ISNSIPIRR716 pKa = 11.84QGRR719 pKa = 11.84HH720 pKa = 3.53WQLMPMSCPNNTRR733 pKa = 11.84GNRR736 pKa = 11.84PRR738 pKa = 11.84QYY740 pKa = 11.05VRR742 pKa = 11.84WW743 pKa = 3.79
Molecular weight: 83.4 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KIH1|A0A1L3KIH1_9VIRU RNA-dependent RNA polymerase OS=Hubei narna-like virus 19 OX=1922949 PE=4 SV=1
MM1 pKa = 6.5TTSSRR6 pKa = 11.84YY7 pKa = 9.24FDD9 pKa = 4.22LPSNILSWPVTPGVVGAGHH28 pKa = 6.25WHH30 pKa = 5.45QLPVSALTDD39 pKa = 3.65RR40 pKa = 11.84DD41 pKa = 3.35GVRR44 pKa = 11.84YY45 pKa = 10.37SNSVWKK51 pKa = 10.52AVVTNIVLAHH61 pKa = 5.85NKK63 pKa = 9.49KK64 pKa = 9.67FRR66 pKa = 11.84NVVPAKK72 pKa = 10.38IFAGWLLRR80 pKa = 11.84CSRR83 pKa = 11.84NAGGRR88 pKa = 11.84YY89 pKa = 8.86YY90 pKa = 10.6VAKK93 pKa = 10.12QLKK96 pKa = 8.74QLSFEE101 pKa = 3.99YY102 pKa = 10.28RR103 pKa = 11.84QWSVTGRR110 pKa = 11.84RR111 pKa = 11.84PTPKK115 pKa = 9.84SDD117 pKa = 3.46VPRR120 pKa = 11.84TFQKK124 pKa = 9.63WLAGHH129 pKa = 5.91VCHH132 pKa = 7.11AEE134 pKa = 3.48ARR136 pKa = 11.84AQIARR141 pKa = 11.84IGRR144 pKa = 11.84CLPEE148 pKa = 5.92GDD150 pKa = 4.26TLVQSKK156 pKa = 10.29AIKK159 pKa = 8.8AHH161 pKa = 6.2RR162 pKa = 11.84SIVTRR167 pKa = 11.84PPCDD171 pKa = 3.42VNMEE175 pKa = 4.25LADD178 pKa = 5.65DD179 pKa = 4.15IYY181 pKa = 10.19YY182 pKa = 9.02WARR185 pKa = 11.84KK186 pKa = 9.02FSTDD190 pKa = 2.95NAHH193 pKa = 6.75LFKK196 pKa = 11.25EE197 pKa = 4.54EE198 pKa = 4.12VSWAPNEE205 pKa = 4.05NSASQGATILEE216 pKa = 4.33GGRR219 pKa = 11.84MEE221 pKa = 5.04EE222 pKa = 4.95LIRR225 pKa = 11.84DD226 pKa = 4.17AKK228 pKa = 11.02AEE230 pKa = 4.04SDD232 pKa = 3.91FEE234 pKa = 4.7SVKK237 pKa = 10.97SLAEE241 pKa = 4.0GDD243 pKa = 3.51FDD245 pKa = 3.86PWVSKK250 pKa = 11.06ALDD253 pKa = 3.59DD254 pKa = 4.11AVYY257 pKa = 7.74MTACTVPPKK266 pKa = 10.51KK267 pKa = 10.22EE268 pKa = 3.99WIPDD272 pKa = 3.35VMAIAEE278 pKa = 4.35KK279 pKa = 10.26GYY281 pKa = 10.07KK282 pKa = 9.95ARR284 pKa = 11.84VLTKK288 pKa = 9.57FPAAALVVGDD298 pKa = 3.76VIRR301 pKa = 11.84RR302 pKa = 11.84QMWAMVSDD310 pKa = 4.36KK311 pKa = 10.41PWADD315 pKa = 3.72MDD317 pKa = 4.04RR318 pKa = 11.84QPDD321 pKa = 3.75DD322 pKa = 3.77VKK324 pKa = 10.88FLSTIRR330 pKa = 11.84SALDD334 pKa = 2.96RR335 pKa = 11.84HH336 pKa = 6.4GICVSSDD343 pKa = 2.8LSNATDD349 pKa = 4.52YY350 pKa = 11.04IPHH353 pKa = 7.33IYY355 pKa = 10.07AQALWAGLLEE365 pKa = 4.31PHH367 pKa = 6.6HH368 pKa = 6.71VMPYY372 pKa = 9.34INSYY376 pKa = 8.58CAIMFSSMKK385 pKa = 10.09LRR387 pKa = 11.84YY388 pKa = 8.99PDD390 pKa = 3.5GTVVTSARR398 pKa = 11.84GIHH401 pKa = 6.19MGTPLSFLTLTLLHH415 pKa = 6.67RR416 pKa = 11.84FCVEE420 pKa = 3.28KK421 pKa = 10.54AGYY424 pKa = 9.96GSYY427 pKa = 10.26PHH429 pKa = 7.66IIRR432 pKa = 11.84GDD434 pKa = 3.67DD435 pKa = 3.45LLGIFPRR442 pKa = 11.84PEE444 pKa = 3.57VYY446 pKa = 10.42FNVMQQVGFSINRR459 pKa = 11.84AKK461 pKa = 10.25TIISRR466 pKa = 11.84TGGTFAEE473 pKa = 4.08RR474 pKa = 11.84TVRR477 pKa = 11.84FSHH480 pKa = 6.75SLATQEE486 pKa = 4.97LSNPLKK492 pKa = 9.94RR493 pKa = 11.84TLGQFIPVNIISSVKK508 pKa = 9.87VLQDD512 pKa = 3.53LPVGGVVRR520 pKa = 11.84ATPGKK525 pKa = 9.75GSLVKK530 pKa = 10.72ALGRR534 pKa = 11.84WFSQTSNIVPRR545 pKa = 11.84QRR547 pKa = 11.84RR548 pKa = 11.84KK549 pKa = 9.53AYY551 pKa = 9.8RR552 pKa = 11.84AIGLNHH558 pKa = 6.97GDD560 pKa = 3.48LVIRR564 pKa = 11.84LSTTVPPHH572 pKa = 6.66LPLDD576 pKa = 4.05LGGAGLPDD584 pKa = 3.09RR585 pKa = 11.84KK586 pKa = 10.62GRR588 pKa = 11.84VGLNGVPFAIRR599 pKa = 11.84AAIGHH604 pKa = 6.53AASHH608 pKa = 6.74HH609 pKa = 4.86EE610 pKa = 4.26TAVKK614 pKa = 9.01LTGLIARR621 pKa = 11.84TDD623 pKa = 3.51GVSRR627 pKa = 11.84GFVDD631 pKa = 3.93VFSKK635 pKa = 10.98KK636 pKa = 10.07DD637 pKa = 3.71FRR639 pKa = 11.84EE640 pKa = 4.04QWSKK644 pKa = 11.35SIWTTEE650 pKa = 3.79PQEE653 pKa = 4.09TEE655 pKa = 4.49FYY657 pKa = 10.71QRR659 pKa = 11.84TRR661 pKa = 11.84RR662 pKa = 11.84YY663 pKa = 7.75WRR665 pKa = 11.84YY666 pKa = 8.76FGHH669 pKa = 7.68RR670 pKa = 11.84DD671 pKa = 3.3RR672 pKa = 11.84PSRR675 pKa = 11.84PISFRR680 pKa = 11.84QWRR683 pKa = 11.84SGLMTLPRR691 pKa = 11.84VKK693 pKa = 10.55ARR695 pKa = 11.84WVPRR699 pKa = 11.84SNSDD703 pKa = 3.12PSRR706 pKa = 11.84LANRR710 pKa = 11.84IRR712 pKa = 11.84SMTGTYY718 pKa = 9.48IPYY721 pKa = 7.82TQPTTGSTYY730 pKa = 9.89QWAAKK735 pKa = 10.17CGG737 pKa = 3.52
MM1 pKa = 6.5TTSSRR6 pKa = 11.84YY7 pKa = 9.24FDD9 pKa = 4.22LPSNILSWPVTPGVVGAGHH28 pKa = 6.25WHH30 pKa = 5.45QLPVSALTDD39 pKa = 3.65RR40 pKa = 11.84DD41 pKa = 3.35GVRR44 pKa = 11.84YY45 pKa = 10.37SNSVWKK51 pKa = 10.52AVVTNIVLAHH61 pKa = 5.85NKK63 pKa = 9.49KK64 pKa = 9.67FRR66 pKa = 11.84NVVPAKK72 pKa = 10.38IFAGWLLRR80 pKa = 11.84CSRR83 pKa = 11.84NAGGRR88 pKa = 11.84YY89 pKa = 8.86YY90 pKa = 10.6VAKK93 pKa = 10.12QLKK96 pKa = 8.74QLSFEE101 pKa = 3.99YY102 pKa = 10.28RR103 pKa = 11.84QWSVTGRR110 pKa = 11.84RR111 pKa = 11.84PTPKK115 pKa = 9.84SDD117 pKa = 3.46VPRR120 pKa = 11.84TFQKK124 pKa = 9.63WLAGHH129 pKa = 5.91VCHH132 pKa = 7.11AEE134 pKa = 3.48ARR136 pKa = 11.84AQIARR141 pKa = 11.84IGRR144 pKa = 11.84CLPEE148 pKa = 5.92GDD150 pKa = 4.26TLVQSKK156 pKa = 10.29AIKK159 pKa = 8.8AHH161 pKa = 6.2RR162 pKa = 11.84SIVTRR167 pKa = 11.84PPCDD171 pKa = 3.42VNMEE175 pKa = 4.25LADD178 pKa = 5.65DD179 pKa = 4.15IYY181 pKa = 10.19YY182 pKa = 9.02WARR185 pKa = 11.84KK186 pKa = 9.02FSTDD190 pKa = 2.95NAHH193 pKa = 6.75LFKK196 pKa = 11.25EE197 pKa = 4.54EE198 pKa = 4.12VSWAPNEE205 pKa = 4.05NSASQGATILEE216 pKa = 4.33GGRR219 pKa = 11.84MEE221 pKa = 5.04EE222 pKa = 4.95LIRR225 pKa = 11.84DD226 pKa = 4.17AKK228 pKa = 11.02AEE230 pKa = 4.04SDD232 pKa = 3.91FEE234 pKa = 4.7SVKK237 pKa = 10.97SLAEE241 pKa = 4.0GDD243 pKa = 3.51FDD245 pKa = 3.86PWVSKK250 pKa = 11.06ALDD253 pKa = 3.59DD254 pKa = 4.11AVYY257 pKa = 7.74MTACTVPPKK266 pKa = 10.51KK267 pKa = 10.22EE268 pKa = 3.99WIPDD272 pKa = 3.35VMAIAEE278 pKa = 4.35KK279 pKa = 10.26GYY281 pKa = 10.07KK282 pKa = 9.95ARR284 pKa = 11.84VLTKK288 pKa = 9.57FPAAALVVGDD298 pKa = 3.76VIRR301 pKa = 11.84RR302 pKa = 11.84QMWAMVSDD310 pKa = 4.36KK311 pKa = 10.41PWADD315 pKa = 3.72MDD317 pKa = 4.04RR318 pKa = 11.84QPDD321 pKa = 3.75DD322 pKa = 3.77VKK324 pKa = 10.88FLSTIRR330 pKa = 11.84SALDD334 pKa = 2.96RR335 pKa = 11.84HH336 pKa = 6.4GICVSSDD343 pKa = 2.8LSNATDD349 pKa = 4.52YY350 pKa = 11.04IPHH353 pKa = 7.33IYY355 pKa = 10.07AQALWAGLLEE365 pKa = 4.31PHH367 pKa = 6.6HH368 pKa = 6.71VMPYY372 pKa = 9.34INSYY376 pKa = 8.58CAIMFSSMKK385 pKa = 10.09LRR387 pKa = 11.84YY388 pKa = 8.99PDD390 pKa = 3.5GTVVTSARR398 pKa = 11.84GIHH401 pKa = 6.19MGTPLSFLTLTLLHH415 pKa = 6.67RR416 pKa = 11.84FCVEE420 pKa = 3.28KK421 pKa = 10.54AGYY424 pKa = 9.96GSYY427 pKa = 10.26PHH429 pKa = 7.66IIRR432 pKa = 11.84GDD434 pKa = 3.67DD435 pKa = 3.45LLGIFPRR442 pKa = 11.84PEE444 pKa = 3.57VYY446 pKa = 10.42FNVMQQVGFSINRR459 pKa = 11.84AKK461 pKa = 10.25TIISRR466 pKa = 11.84TGGTFAEE473 pKa = 4.08RR474 pKa = 11.84TVRR477 pKa = 11.84FSHH480 pKa = 6.75SLATQEE486 pKa = 4.97LSNPLKK492 pKa = 9.94RR493 pKa = 11.84TLGQFIPVNIISSVKK508 pKa = 9.87VLQDD512 pKa = 3.53LPVGGVVRR520 pKa = 11.84ATPGKK525 pKa = 9.75GSLVKK530 pKa = 10.72ALGRR534 pKa = 11.84WFSQTSNIVPRR545 pKa = 11.84QRR547 pKa = 11.84RR548 pKa = 11.84KK549 pKa = 9.53AYY551 pKa = 9.8RR552 pKa = 11.84AIGLNHH558 pKa = 6.97GDD560 pKa = 3.48LVIRR564 pKa = 11.84LSTTVPPHH572 pKa = 6.66LPLDD576 pKa = 4.05LGGAGLPDD584 pKa = 3.09RR585 pKa = 11.84KK586 pKa = 10.62GRR588 pKa = 11.84VGLNGVPFAIRR599 pKa = 11.84AAIGHH604 pKa = 6.53AASHH608 pKa = 6.74HH609 pKa = 4.86EE610 pKa = 4.26TAVKK614 pKa = 9.01LTGLIARR621 pKa = 11.84TDD623 pKa = 3.51GVSRR627 pKa = 11.84GFVDD631 pKa = 3.93VFSKK635 pKa = 10.98KK636 pKa = 10.07DD637 pKa = 3.71FRR639 pKa = 11.84EE640 pKa = 4.04QWSKK644 pKa = 11.35SIWTTEE650 pKa = 3.79PQEE653 pKa = 4.09TEE655 pKa = 4.49FYY657 pKa = 10.71QRR659 pKa = 11.84TRR661 pKa = 11.84RR662 pKa = 11.84YY663 pKa = 7.75WRR665 pKa = 11.84YY666 pKa = 8.76FGHH669 pKa = 7.68RR670 pKa = 11.84DD671 pKa = 3.3RR672 pKa = 11.84PSRR675 pKa = 11.84PISFRR680 pKa = 11.84QWRR683 pKa = 11.84SGLMTLPRR691 pKa = 11.84VKK693 pKa = 10.55ARR695 pKa = 11.84WVPRR699 pKa = 11.84SNSDD703 pKa = 3.12PSRR706 pKa = 11.84LANRR710 pKa = 11.84IRR712 pKa = 11.84SMTGTYY718 pKa = 9.48IPYY721 pKa = 7.82TQPTTGSTYY730 pKa = 9.89QWAAKK735 pKa = 10.17CGG737 pKa = 3.52
Molecular weight: 82.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1480 |
737 |
743 |
740.0 |
83.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.824 ± 1.111 | 2.162 ± 0.658 |
4.662 ± 0.251 | 4.662 ± 0.888 |
3.243 ± 0.389 | 7.432 ± 0.264 |
3.446 ± 0.417 | 5.27 ± 0.015 |
3.986 ± 0.723 | 8.243 ± 0.546 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.824 ± 0.148 | 3.581 ± 0.512 |
6.149 ± 0.03 | 2.838 ± 0.293 |
9.392 ± 0.59 | 7.973 ± 0.167 |
5.878 ± 0.254 | 7.5 ± 0.164 |
2.162 ± 0.386 | 2.77 ± 0.34 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
