
Fall chinook aquareovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Spinareovirinae; Aquareovirus; unclassified Aquareovirus
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
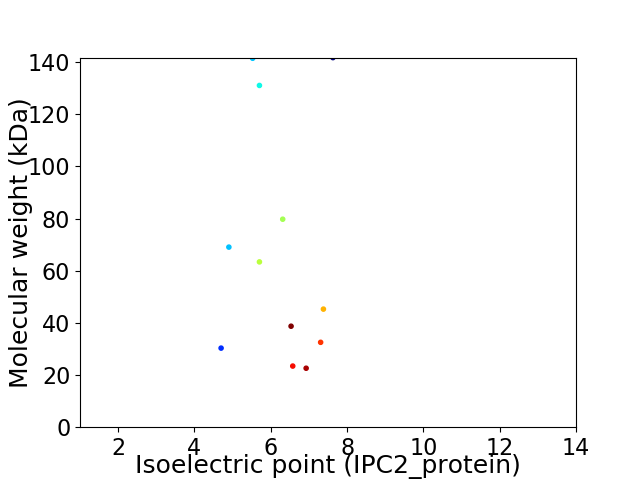
Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1S6PCW9|A0A1S6PCW9_9REOV NS4 OS=Fall chinook aquareovirus OX=1963254 PE=4 SV=1
MM1 pKa = 7.25EE2 pKa = 5.66FKK4 pKa = 10.62RR5 pKa = 11.84GVPTVSVHH13 pKa = 7.23RR14 pKa = 11.84DD15 pKa = 3.02MTEE18 pKa = 3.89FPEE21 pKa = 4.64VATPPIDD28 pKa = 5.4AIFDD32 pKa = 3.61QAQPYY37 pKa = 9.35CDD39 pKa = 4.32HH40 pKa = 7.04LGKK43 pKa = 9.76PLISVYY49 pKa = 10.55DD50 pKa = 3.67GRR52 pKa = 11.84LVPISGPTHH61 pKa = 7.2LEE63 pKa = 4.13DD64 pKa = 4.64EE65 pKa = 4.99DD66 pKa = 6.23DD67 pKa = 3.98YY68 pKa = 11.94EE69 pKa = 4.23PWQVHH74 pKa = 5.09GQLQTLSAHH83 pKa = 5.88SKK85 pKa = 9.04WPEE88 pKa = 3.91SVPNWTSHH96 pKa = 4.76PHH98 pKa = 5.02IQVFSNMSPYY108 pKa = 11.34ANIPGPSPYY117 pKa = 10.34KK118 pKa = 10.38FEE120 pKa = 4.2SWPAALAPHH129 pKa = 6.99GGLSAWDD136 pKa = 4.05LLWRR140 pKa = 11.84CAQLRR145 pKa = 11.84PNRR148 pKa = 11.84LPVYY152 pKa = 10.41SDD154 pKa = 3.79AFLLSTLRR162 pKa = 11.84LSGKK166 pKa = 8.05QVFEE170 pKa = 4.52LPMLDD175 pKa = 3.69SVADD179 pKa = 3.78MLSWAGWNADD189 pKa = 3.86MLCGLDD195 pKa = 3.54LCQTEE200 pKa = 4.26GMMAIPDD207 pKa = 4.56PGDD210 pKa = 3.12EE211 pKa = 4.33HH212 pKa = 8.32LPLLVEE218 pKa = 3.73QTAWFDD224 pKa = 3.75YY225 pKa = 10.5IIRR228 pKa = 11.84LPFDD232 pKa = 3.62TTHH235 pKa = 6.77HH236 pKa = 6.72LGLGLTDD243 pKa = 3.89DD244 pKa = 4.76QLWSHH249 pKa = 7.52PFTVLCFLRR258 pKa = 11.84WRR260 pKa = 11.84AAWPDD265 pKa = 3.72LYY267 pKa = 11.45
MM1 pKa = 7.25EE2 pKa = 5.66FKK4 pKa = 10.62RR5 pKa = 11.84GVPTVSVHH13 pKa = 7.23RR14 pKa = 11.84DD15 pKa = 3.02MTEE18 pKa = 3.89FPEE21 pKa = 4.64VATPPIDD28 pKa = 5.4AIFDD32 pKa = 3.61QAQPYY37 pKa = 9.35CDD39 pKa = 4.32HH40 pKa = 7.04LGKK43 pKa = 9.76PLISVYY49 pKa = 10.55DD50 pKa = 3.67GRR52 pKa = 11.84LVPISGPTHH61 pKa = 7.2LEE63 pKa = 4.13DD64 pKa = 4.64EE65 pKa = 4.99DD66 pKa = 6.23DD67 pKa = 3.98YY68 pKa = 11.94EE69 pKa = 4.23PWQVHH74 pKa = 5.09GQLQTLSAHH83 pKa = 5.88SKK85 pKa = 9.04WPEE88 pKa = 3.91SVPNWTSHH96 pKa = 4.76PHH98 pKa = 5.02IQVFSNMSPYY108 pKa = 11.34ANIPGPSPYY117 pKa = 10.34KK118 pKa = 10.38FEE120 pKa = 4.2SWPAALAPHH129 pKa = 6.99GGLSAWDD136 pKa = 4.05LLWRR140 pKa = 11.84CAQLRR145 pKa = 11.84PNRR148 pKa = 11.84LPVYY152 pKa = 10.41SDD154 pKa = 3.79AFLLSTLRR162 pKa = 11.84LSGKK166 pKa = 8.05QVFEE170 pKa = 4.52LPMLDD175 pKa = 3.69SVADD179 pKa = 3.78MLSWAGWNADD189 pKa = 3.86MLCGLDD195 pKa = 3.54LCQTEE200 pKa = 4.26GMMAIPDD207 pKa = 4.56PGDD210 pKa = 3.12EE211 pKa = 4.33HH212 pKa = 8.32LPLLVEE218 pKa = 3.73QTAWFDD224 pKa = 3.75YY225 pKa = 10.5IIRR228 pKa = 11.84LPFDD232 pKa = 3.62TTHH235 pKa = 6.77HH236 pKa = 6.72LGLGLTDD243 pKa = 3.89DD244 pKa = 4.76QLWSHH249 pKa = 7.52PFTVLCFLRR258 pKa = 11.84WRR260 pKa = 11.84AAWPDD265 pKa = 3.72LYY267 pKa = 11.45
Molecular weight: 30.32 kDa
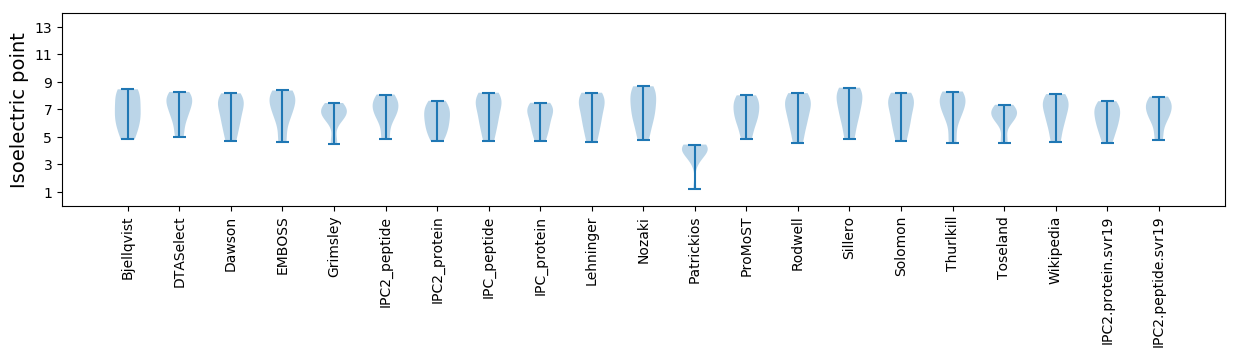
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1S6PCW6|A0A1S6PCW6_9REOV NS2 OS=Fall chinook aquareovirus OX=1963254 PE=4 SV=1
MM1 pKa = 7.83PCGDD5 pKa = 4.48TIATTFQYY13 pKa = 10.51TVLQVDD19 pKa = 4.62RR20 pKa = 11.84SCCISTSVTATSEE33 pKa = 4.11ATSWALPPILACCCCVCCCAAAGYY57 pKa = 9.91AISRR61 pKa = 11.84GSCPCITRR69 pKa = 11.84RR70 pKa = 11.84LDD72 pKa = 3.18VLRR75 pKa = 11.84GTGFTTEE82 pKa = 3.48HH83 pKa = 6.57SLRR86 pKa = 11.84ANRR89 pKa = 11.84KK90 pKa = 8.2SISRR94 pKa = 11.84SHH96 pKa = 6.93HH97 pKa = 5.8SRR99 pKa = 11.84SSDD102 pKa = 3.2SRR104 pKa = 11.84STSHH108 pKa = 6.97HH109 pKa = 6.83PALEE113 pKa = 4.27LPGYY117 pKa = 10.18RR118 pKa = 11.84YY119 pKa = 8.51TDD121 pKa = 2.71VSYY124 pKa = 11.31AHH126 pKa = 7.21RR127 pKa = 11.84SEE129 pKa = 4.39SDD131 pKa = 3.35SASEE135 pKa = 3.9LSSAEE140 pKa = 4.16SEE142 pKa = 4.56VITYY146 pKa = 7.5QPRR149 pKa = 11.84PFITHH154 pKa = 7.13LEE156 pKa = 4.26SQSSCGTCLDD166 pKa = 3.82HH167 pKa = 6.33QQNGIQEE174 pKa = 4.38RR175 pKa = 11.84SPNCVSPSRR184 pKa = 11.84HH185 pKa = 5.61DD186 pKa = 4.15RR187 pKa = 11.84IPRR190 pKa = 11.84SGHH193 pKa = 4.04PTYY196 pKa = 10.81RR197 pKa = 11.84CDD199 pKa = 4.09LRR201 pKa = 11.84PGSTLLL207 pKa = 4.01
MM1 pKa = 7.83PCGDD5 pKa = 4.48TIATTFQYY13 pKa = 10.51TVLQVDD19 pKa = 4.62RR20 pKa = 11.84SCCISTSVTATSEE33 pKa = 4.11ATSWALPPILACCCCVCCCAAAGYY57 pKa = 9.91AISRR61 pKa = 11.84GSCPCITRR69 pKa = 11.84RR70 pKa = 11.84LDD72 pKa = 3.18VLRR75 pKa = 11.84GTGFTTEE82 pKa = 3.48HH83 pKa = 6.57SLRR86 pKa = 11.84ANRR89 pKa = 11.84KK90 pKa = 8.2SISRR94 pKa = 11.84SHH96 pKa = 6.93HH97 pKa = 5.8SRR99 pKa = 11.84SSDD102 pKa = 3.2SRR104 pKa = 11.84STSHH108 pKa = 6.97HH109 pKa = 6.83PALEE113 pKa = 4.27LPGYY117 pKa = 10.18RR118 pKa = 11.84YY119 pKa = 8.51TDD121 pKa = 2.71VSYY124 pKa = 11.31AHH126 pKa = 7.21RR127 pKa = 11.84SEE129 pKa = 4.39SDD131 pKa = 3.35SASEE135 pKa = 3.9LSSAEE140 pKa = 4.16SEE142 pKa = 4.56VITYY146 pKa = 7.5QPRR149 pKa = 11.84PFITHH154 pKa = 7.13LEE156 pKa = 4.26SQSSCGTCLDD166 pKa = 3.82HH167 pKa = 6.33QQNGIQEE174 pKa = 4.38RR175 pKa = 11.84SPNCVSPSRR184 pKa = 11.84HH185 pKa = 5.61DD186 pKa = 4.15RR187 pKa = 11.84IPRR190 pKa = 11.84SGHH193 pKa = 4.04PTYY196 pKa = 10.81RR197 pKa = 11.84CDD199 pKa = 4.09LRR201 pKa = 11.84PGSTLLL207 pKa = 4.01
Molecular weight: 22.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7502 |
207 |
1295 |
625.2 |
68.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.797 ± 0.468 | 1.293 ± 0.271 |
5.319 ± 0.226 | 3.159 ± 0.231 |
3.479 ± 0.244 | 5.678 ± 0.216 |
2.706 ± 0.261 | 5.385 ± 0.251 |
2.319 ± 0.347 | 9.717 ± 0.439 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.813 ± 0.268 | 4.132 ± 0.275 |
7.665 ± 0.348 | 3.759 ± 0.292 |
5.305 ± 0.319 | 8.438 ± 0.572 |
7.305 ± 0.35 | 7.265 ± 0.318 |
1.533 ± 0.218 | 2.933 ± 0.251 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
