
Pistricoccus aurantiacus
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Oceanospirillales; Halomonadaceae; Pistricoccus
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins
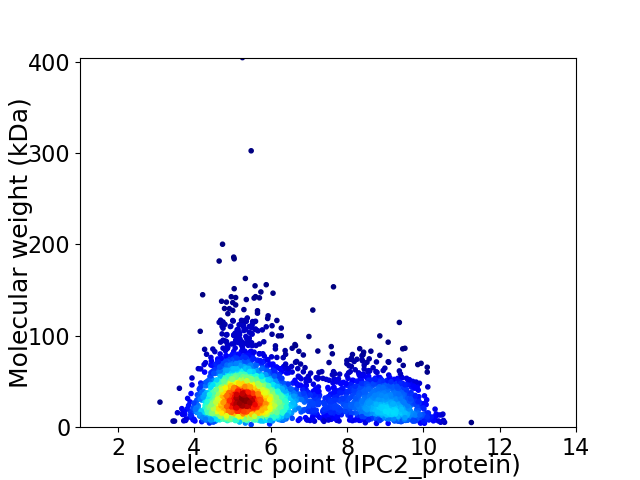
Virtual 2D-PAGE plot for 3396 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5B8SWT2|A0A5B8SWT2_9GAMM Respiratory nitrate reductase subunit beta OS=Pistricoccus aurantiacus OX=1883414 GN=FGL86_17045 PE=4 SV=1
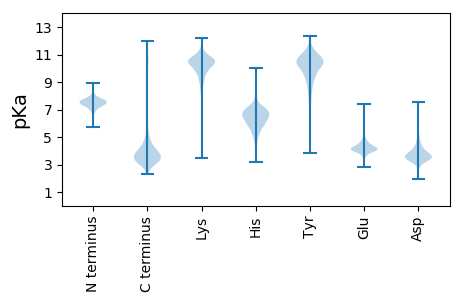
MM1 pKa = 7.72KK2 pKa = 9.1KK3 pKa = 8.18TLLVTAIAGALAASAGAASAATVYY27 pKa = 10.76NQDD30 pKa = 3.0GTKK33 pKa = 10.37LDD35 pKa = 3.96LYY37 pKa = 11.59GNIQLAYY44 pKa = 9.77KK45 pKa = 10.32SIEE48 pKa = 3.92TAQFEE53 pKa = 4.54DD54 pKa = 4.08VPGVGNDD61 pKa = 2.85IYY63 pKa = 11.0QGTEE67 pKa = 3.7AEE69 pKa = 5.87DD70 pKa = 4.09EE71 pKa = 4.25IFDD74 pKa = 3.91NGSTIGVTGEE84 pKa = 3.86HH85 pKa = 6.64VIMNGLTGYY94 pKa = 10.16FKK96 pKa = 11.28AEE98 pKa = 3.84WEE100 pKa = 4.13FDD102 pKa = 3.07ADD104 pKa = 3.83QAKK107 pKa = 9.67GAPGGINTGDD117 pKa = 3.29QAYY120 pKa = 10.61LGLKK124 pKa = 10.26GNFGDD129 pKa = 4.77ARR131 pKa = 11.84LGSWDD136 pKa = 4.23PLIDD140 pKa = 5.15DD141 pKa = 5.78WIQDD145 pKa = 3.91PISNNEE151 pKa = 3.8YY152 pKa = 10.85FDD154 pKa = 4.5VSDD157 pKa = 3.66SNGRR161 pKa = 11.84ILGVEE166 pKa = 3.73NRR168 pKa = 11.84EE169 pKa = 3.98GDD171 pKa = 3.4KK172 pKa = 10.64LQYY175 pKa = 9.38MSPSLGGLQFAVGTQYY191 pKa = 11.35KK192 pKa = 10.49GDD194 pKa = 3.89AEE196 pKa = 4.28FDD198 pKa = 3.55GDD200 pKa = 4.16DD201 pKa = 3.94TVTGTNSLGDD211 pKa = 4.23EE212 pKa = 4.11ITVTGEE218 pKa = 3.88DD219 pKa = 3.29SSNASFFGGLKK230 pKa = 10.2YY231 pKa = 10.46EE232 pKa = 4.19VGNFSIAAVYY242 pKa = 10.87DD243 pKa = 3.62NLDD246 pKa = 3.95NNDD249 pKa = 3.34GSYY252 pKa = 10.95SAVDD256 pKa = 3.24SDD258 pKa = 4.74GNDD261 pKa = 2.93IGSGDD266 pKa = 3.85FEE268 pKa = 6.42AGEE271 pKa = 4.32QYY273 pKa = 10.94GITGQYY279 pKa = 8.27TWDD282 pKa = 3.69TLRR285 pKa = 11.84VALKK289 pKa = 10.78AEE291 pKa = 4.31RR292 pKa = 11.84FKK294 pKa = 11.52SDD296 pKa = 3.37NDD298 pKa = 3.7TLADD302 pKa = 3.54TNYY305 pKa = 8.8YY306 pKa = 10.89ALGARR311 pKa = 11.84YY312 pKa = 9.63GYY314 pKa = 10.37GNGMGDD320 pKa = 4.06LYY322 pKa = 11.41GSYY325 pKa = 10.49QYY327 pKa = 11.58VDD329 pKa = 3.21VGGGDD334 pKa = 4.06FLDD337 pKa = 4.24TADD340 pKa = 4.96DD341 pKa = 4.34ALTSGDD347 pKa = 3.49WPSEE351 pKa = 4.17RR352 pKa = 11.84EE353 pKa = 3.9DD354 pKa = 3.44EE355 pKa = 4.49SYY357 pKa = 11.75NEE359 pKa = 4.51IILGATYY366 pKa = 10.51NISDD370 pKa = 3.26AMYY373 pKa = 9.27TFVEE377 pKa = 4.5GALYY381 pKa = 10.68DD382 pKa = 3.78RR383 pKa = 11.84TDD385 pKa = 3.62DD386 pKa = 3.94EE387 pKa = 6.37GDD389 pKa = 3.61GVAVGAVYY397 pKa = 10.44LFF399 pKa = 4.21
MM1 pKa = 7.72KK2 pKa = 9.1KK3 pKa = 8.18TLLVTAIAGALAASAGAASAATVYY27 pKa = 10.76NQDD30 pKa = 3.0GTKK33 pKa = 10.37LDD35 pKa = 3.96LYY37 pKa = 11.59GNIQLAYY44 pKa = 9.77KK45 pKa = 10.32SIEE48 pKa = 3.92TAQFEE53 pKa = 4.54DD54 pKa = 4.08VPGVGNDD61 pKa = 2.85IYY63 pKa = 11.0QGTEE67 pKa = 3.7AEE69 pKa = 5.87DD70 pKa = 4.09EE71 pKa = 4.25IFDD74 pKa = 3.91NGSTIGVTGEE84 pKa = 3.86HH85 pKa = 6.64VIMNGLTGYY94 pKa = 10.16FKK96 pKa = 11.28AEE98 pKa = 3.84WEE100 pKa = 4.13FDD102 pKa = 3.07ADD104 pKa = 3.83QAKK107 pKa = 9.67GAPGGINTGDD117 pKa = 3.29QAYY120 pKa = 10.61LGLKK124 pKa = 10.26GNFGDD129 pKa = 4.77ARR131 pKa = 11.84LGSWDD136 pKa = 4.23PLIDD140 pKa = 5.15DD141 pKa = 5.78WIQDD145 pKa = 3.91PISNNEE151 pKa = 3.8YY152 pKa = 10.85FDD154 pKa = 4.5VSDD157 pKa = 3.66SNGRR161 pKa = 11.84ILGVEE166 pKa = 3.73NRR168 pKa = 11.84EE169 pKa = 3.98GDD171 pKa = 3.4KK172 pKa = 10.64LQYY175 pKa = 9.38MSPSLGGLQFAVGTQYY191 pKa = 11.35KK192 pKa = 10.49GDD194 pKa = 3.89AEE196 pKa = 4.28FDD198 pKa = 3.55GDD200 pKa = 4.16DD201 pKa = 3.94TVTGTNSLGDD211 pKa = 4.23EE212 pKa = 4.11ITVTGEE218 pKa = 3.88DD219 pKa = 3.29SSNASFFGGLKK230 pKa = 10.2YY231 pKa = 10.46EE232 pKa = 4.19VGNFSIAAVYY242 pKa = 10.87DD243 pKa = 3.62NLDD246 pKa = 3.95NNDD249 pKa = 3.34GSYY252 pKa = 10.95SAVDD256 pKa = 3.24SDD258 pKa = 4.74GNDD261 pKa = 2.93IGSGDD266 pKa = 3.85FEE268 pKa = 6.42AGEE271 pKa = 4.32QYY273 pKa = 10.94GITGQYY279 pKa = 8.27TWDD282 pKa = 3.69TLRR285 pKa = 11.84VALKK289 pKa = 10.78AEE291 pKa = 4.31RR292 pKa = 11.84FKK294 pKa = 11.52SDD296 pKa = 3.37NDD298 pKa = 3.7TLADD302 pKa = 3.54TNYY305 pKa = 8.8YY306 pKa = 10.89ALGARR311 pKa = 11.84YY312 pKa = 9.63GYY314 pKa = 10.37GNGMGDD320 pKa = 4.06LYY322 pKa = 11.41GSYY325 pKa = 10.49QYY327 pKa = 11.58VDD329 pKa = 3.21VGGGDD334 pKa = 4.06FLDD337 pKa = 4.24TADD340 pKa = 4.96DD341 pKa = 4.34ALTSGDD347 pKa = 3.49WPSEE351 pKa = 4.17RR352 pKa = 11.84EE353 pKa = 3.9DD354 pKa = 3.44EE355 pKa = 4.49SYY357 pKa = 11.75NEE359 pKa = 4.51IILGATYY366 pKa = 10.51NISDD370 pKa = 3.26AMYY373 pKa = 9.27TFVEE377 pKa = 4.5GALYY381 pKa = 10.68DD382 pKa = 3.78RR383 pKa = 11.84TDD385 pKa = 3.62DD386 pKa = 3.94EE387 pKa = 6.37GDD389 pKa = 3.61GVAVGAVYY397 pKa = 10.44LFF399 pKa = 4.21
Molecular weight: 42.61 kDa
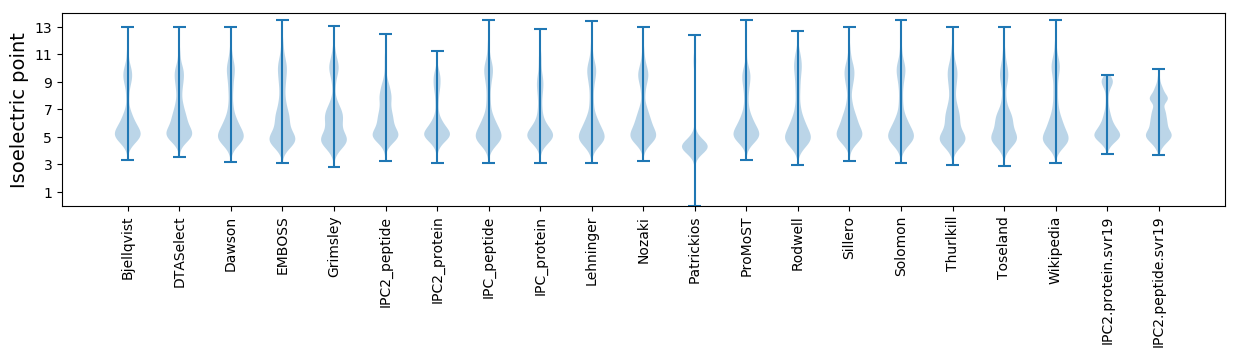
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5B8SNT8|A0A5B8SNT8_9GAMM 30S ribosomal protein S20 OS=Pistricoccus aurantiacus OX=1883414 GN=rpsT PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVIKK11 pKa = 10.38RR12 pKa = 11.84KK13 pKa = 9.23RR14 pKa = 11.84VHH16 pKa = 6.26GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.3NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVIKK11 pKa = 10.38RR12 pKa = 11.84KK13 pKa = 9.23RR14 pKa = 11.84VHH16 pKa = 6.26GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.3NGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1106925 |
28 |
3619 |
325.9 |
36.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.487 ± 0.046 | 0.945 ± 0.013 |
5.708 ± 0.038 | 6.497 ± 0.043 |
3.597 ± 0.024 | 7.881 ± 0.032 |
2.307 ± 0.022 | 5.089 ± 0.035 |
3.226 ± 0.032 | 11.522 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.449 ± 0.02 | 2.778 ± 0.02 |
4.868 ± 0.026 | 4.097 ± 0.03 |
7.17 ± 0.041 | 5.578 ± 0.025 |
4.926 ± 0.025 | 6.822 ± 0.035 |
1.549 ± 0.019 | 2.505 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
