
Clostridium phage JD032
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; Sherbrookevirus; unclassified Sherbrookevirus
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
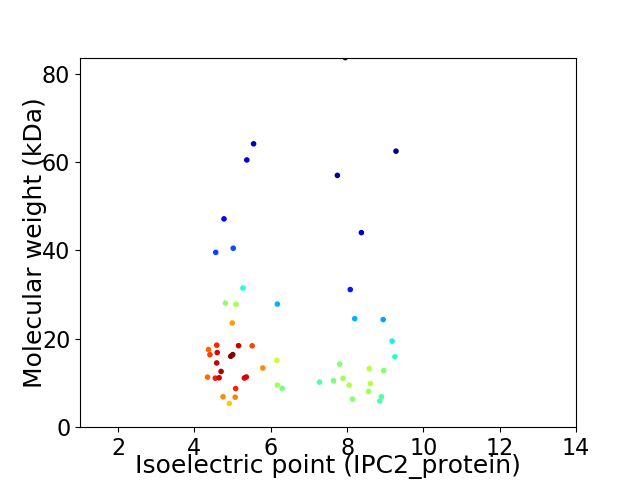
Virtual 2D-PAGE plot for 54 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6B7FSJ2|A0A6B7FSJ2_9CAUD Baseplate protein J OS=Clostridium phage JD032 OX=2530171 GN=JD032_36 PE=4 SV=1
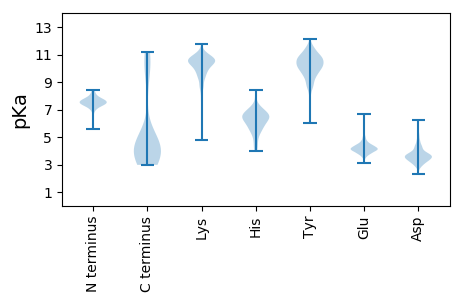
MM1 pKa = 7.63EE2 pKa = 4.89KK3 pKa = 10.3FIRR6 pKa = 11.84LDD8 pKa = 3.23YY9 pKa = 11.45DD10 pKa = 3.04KK11 pKa = 11.36GFRR14 pKa = 11.84GKK16 pKa = 9.75EE17 pKa = 4.03HH18 pKa = 6.43VSSATGDD25 pKa = 3.82GEE27 pKa = 4.23HH28 pKa = 6.7FEE30 pKa = 5.39AGISCYY36 pKa = 10.24KK37 pKa = 9.75ISKK40 pKa = 7.78EE41 pKa = 3.95KK42 pKa = 10.98CVDD45 pKa = 4.07AIINLCEE52 pKa = 3.76YY53 pKa = 8.8WFEE56 pKa = 4.51FAGEE60 pKa = 4.23CQFKK64 pKa = 11.12DD65 pKa = 3.22FDD67 pKa = 4.35INIFEE72 pKa = 4.33GCYY75 pKa = 9.26VGEE78 pKa = 4.31GASYY82 pKa = 11.0EE83 pKa = 4.15DD84 pKa = 4.32LATCEE89 pKa = 3.87KK90 pKa = 10.63HH91 pKa = 6.7LYY93 pKa = 8.5TVDD96 pKa = 3.22GSLFNEE102 pKa = 4.76VYY104 pKa = 10.54DD105 pKa = 4.23LYY107 pKa = 11.56YY108 pKa = 9.88MHH110 pKa = 6.84EE111 pKa = 4.48TYY113 pKa = 10.81LEE115 pKa = 4.05EE116 pKa = 4.29NGNVEE121 pKa = 4.3EE122 pKa = 5.38LEE124 pKa = 4.12EE125 pKa = 4.2NYY127 pKa = 10.19KK128 pKa = 10.86DD129 pKa = 3.78EE130 pKa = 5.0YY131 pKa = 10.02ITTEE135 pKa = 3.8EE136 pKa = 4.41FEE138 pKa = 4.55TKK140 pKa = 9.71IKK142 pKa = 10.83EE143 pKa = 3.99MFIKK147 pKa = 10.79YY148 pKa = 9.62LL149 pKa = 3.6
MM1 pKa = 7.63EE2 pKa = 4.89KK3 pKa = 10.3FIRR6 pKa = 11.84LDD8 pKa = 3.23YY9 pKa = 11.45DD10 pKa = 3.04KK11 pKa = 11.36GFRR14 pKa = 11.84GKK16 pKa = 9.75EE17 pKa = 4.03HH18 pKa = 6.43VSSATGDD25 pKa = 3.82GEE27 pKa = 4.23HH28 pKa = 6.7FEE30 pKa = 5.39AGISCYY36 pKa = 10.24KK37 pKa = 9.75ISKK40 pKa = 7.78EE41 pKa = 3.95KK42 pKa = 10.98CVDD45 pKa = 4.07AIINLCEE52 pKa = 3.76YY53 pKa = 8.8WFEE56 pKa = 4.51FAGEE60 pKa = 4.23CQFKK64 pKa = 11.12DD65 pKa = 3.22FDD67 pKa = 4.35INIFEE72 pKa = 4.33GCYY75 pKa = 9.26VGEE78 pKa = 4.31GASYY82 pKa = 11.0EE83 pKa = 4.15DD84 pKa = 4.32LATCEE89 pKa = 3.87KK90 pKa = 10.63HH91 pKa = 6.7LYY93 pKa = 8.5TVDD96 pKa = 3.22GSLFNEE102 pKa = 4.76VYY104 pKa = 10.54DD105 pKa = 4.23LYY107 pKa = 11.56YY108 pKa = 9.88MHH110 pKa = 6.84EE111 pKa = 4.48TYY113 pKa = 10.81LEE115 pKa = 4.05EE116 pKa = 4.29NGNVEE121 pKa = 4.3EE122 pKa = 5.38LEE124 pKa = 4.12EE125 pKa = 4.2NYY127 pKa = 10.19KK128 pKa = 10.86DD129 pKa = 3.78EE130 pKa = 5.0YY131 pKa = 10.02ITTEE135 pKa = 3.8EE136 pKa = 4.41FEE138 pKa = 4.55TKK140 pKa = 9.71IKK142 pKa = 10.83EE143 pKa = 3.99MFIKK147 pKa = 10.79YY148 pKa = 9.62LL149 pKa = 3.6
Molecular weight: 17.58 kDa
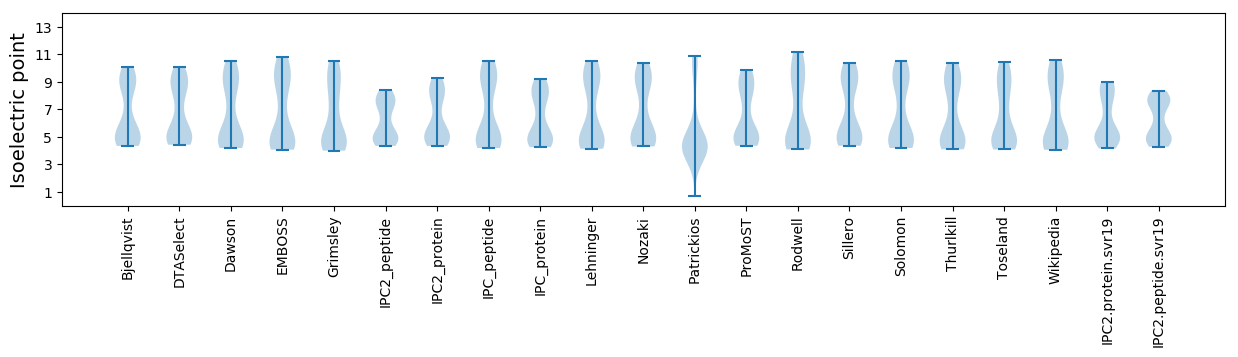
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6B7FUN9|A0A6B7FUN9_9CAUD ATP-dependent Clp protease proteolytic subunit OS=Clostridium phage JD032 OX=2530171 GN=JD032_51 PE=3 SV=1
MM1 pKa = 7.5ARR3 pKa = 11.84RR4 pKa = 11.84HH5 pKa = 5.35IGAVISLKK13 pKa = 11.11DD14 pKa = 3.63NMSATMRR21 pKa = 11.84GIRR24 pKa = 11.84RR25 pKa = 11.84EE26 pKa = 3.74QKK28 pKa = 9.29QFQNEE33 pKa = 3.76VRR35 pKa = 11.84RR36 pKa = 11.84TRR38 pKa = 11.84NEE40 pKa = 3.3MRR42 pKa = 11.84SASRR46 pKa = 11.84EE47 pKa = 3.67RR48 pKa = 11.84MRR50 pKa = 11.84IRR52 pKa = 11.84MDD54 pKa = 3.02ATPAHH59 pKa = 6.58RR60 pKa = 11.84TIQDD64 pKa = 3.4LRR66 pKa = 11.84RR67 pKa = 11.84KK68 pKa = 8.49FAPLRR73 pKa = 11.84TKK75 pKa = 10.2LVKK78 pKa = 10.52AVVIKK83 pKa = 10.82DD84 pKa = 3.35LATEE88 pKa = 4.19KK89 pKa = 10.45IEE91 pKa = 4.58RR92 pKa = 11.84IKK94 pKa = 11.47SNVKK98 pKa = 10.41SFGKK102 pKa = 10.36FIARR106 pKa = 11.84PVVKK110 pKa = 10.65LKK112 pKa = 11.13DD113 pKa = 3.56EE114 pKa = 4.36TKK116 pKa = 11.22GMIDD120 pKa = 3.48KK121 pKa = 10.47IKK123 pKa = 10.98NRR125 pKa = 11.84LTSLSTIVPVGAAVGAAGMAVKK147 pKa = 10.35SGMEE151 pKa = 4.22LEE153 pKa = 4.18QQQISMRR160 pKa = 11.84HH161 pKa = 4.12FMGVGNKK168 pKa = 9.42GKK170 pKa = 10.31SSKK173 pKa = 10.53EE174 pKa = 3.68LDD176 pKa = 3.7GMSASYY182 pKa = 11.0LKK184 pKa = 10.53DD185 pKa = 3.17LRR187 pKa = 11.84NNANATPFEE196 pKa = 4.2TGEE199 pKa = 4.29VISAGTRR206 pKa = 11.84SLQIAGGNTKK216 pKa = 10.71DD217 pKa = 3.26AMQMVKK223 pKa = 10.34LAEE226 pKa = 4.7DD227 pKa = 3.64MAALNPGKK235 pKa = 8.56TVGDD239 pKa = 3.67AMEE242 pKa = 5.22ALADD246 pKa = 3.83MNIGEE251 pKa = 4.53MARR254 pKa = 11.84LTEE257 pKa = 4.43FGVKK261 pKa = 10.25ASSTDD266 pKa = 3.47DD267 pKa = 3.52PKK269 pKa = 11.15EE270 pKa = 3.99VQKK273 pKa = 11.17KK274 pKa = 10.2LEE276 pKa = 3.87TMYY279 pKa = 11.28AGGANKK285 pKa = 9.88LAEE288 pKa = 4.68SGSGLLSTIMGKK300 pKa = 10.14LKK302 pKa = 11.12SNIADD307 pKa = 3.2IGLGMLEE314 pKa = 4.03PLKK317 pKa = 10.63PVMTGLIGFIDD328 pKa = 3.69QASPKK333 pKa = 9.45ILEE336 pKa = 4.37VGTKK340 pKa = 7.49ITSGIGMAIGWIQQQMPTLAPIFQTAFGAVSSIVSTVAPIIGQVIGALSPVFMGLLSVASSALSGIASAVKK411 pKa = 8.64TVAPVVSSLISKK423 pKa = 7.97LAPIFLNVGSTLKK436 pKa = 11.14SMGKK440 pKa = 9.07IFKK443 pKa = 10.37NVFDD447 pKa = 4.39SVMKK451 pKa = 10.19IVKK454 pKa = 9.55KK455 pKa = 10.7ASDD458 pKa = 4.07FIKK461 pKa = 10.43PLVNGIIGATKK472 pKa = 10.29GISDD476 pKa = 3.93GVSWVAGKK484 pKa = 9.91LAGNATGTKK493 pKa = 8.26YY494 pKa = 10.08WSGGLSVVGEE504 pKa = 4.49HH505 pKa = 6.58GPEE508 pKa = 3.95LVSMPRR514 pKa = 11.84GSKK517 pKa = 10.23VFTNAEE523 pKa = 4.23SKK525 pKa = 11.46SMINKK530 pKa = 9.38SIPNFRR536 pKa = 11.84QVQGEE541 pKa = 4.12NTNYY545 pKa = 10.71NITIPKK551 pKa = 9.09IAEE554 pKa = 3.91QIVIRR559 pKa = 11.84EE560 pKa = 4.05DD561 pKa = 3.24ADD563 pKa = 3.29IEE565 pKa = 4.78RR566 pKa = 11.84ITSSLIKK573 pKa = 9.61KK574 pKa = 9.18IQMAKK579 pKa = 9.32MGGVVV584 pKa = 2.99
MM1 pKa = 7.5ARR3 pKa = 11.84RR4 pKa = 11.84HH5 pKa = 5.35IGAVISLKK13 pKa = 11.11DD14 pKa = 3.63NMSATMRR21 pKa = 11.84GIRR24 pKa = 11.84RR25 pKa = 11.84EE26 pKa = 3.74QKK28 pKa = 9.29QFQNEE33 pKa = 3.76VRR35 pKa = 11.84RR36 pKa = 11.84TRR38 pKa = 11.84NEE40 pKa = 3.3MRR42 pKa = 11.84SASRR46 pKa = 11.84EE47 pKa = 3.67RR48 pKa = 11.84MRR50 pKa = 11.84IRR52 pKa = 11.84MDD54 pKa = 3.02ATPAHH59 pKa = 6.58RR60 pKa = 11.84TIQDD64 pKa = 3.4LRR66 pKa = 11.84RR67 pKa = 11.84KK68 pKa = 8.49FAPLRR73 pKa = 11.84TKK75 pKa = 10.2LVKK78 pKa = 10.52AVVIKK83 pKa = 10.82DD84 pKa = 3.35LATEE88 pKa = 4.19KK89 pKa = 10.45IEE91 pKa = 4.58RR92 pKa = 11.84IKK94 pKa = 11.47SNVKK98 pKa = 10.41SFGKK102 pKa = 10.36FIARR106 pKa = 11.84PVVKK110 pKa = 10.65LKK112 pKa = 11.13DD113 pKa = 3.56EE114 pKa = 4.36TKK116 pKa = 11.22GMIDD120 pKa = 3.48KK121 pKa = 10.47IKK123 pKa = 10.98NRR125 pKa = 11.84LTSLSTIVPVGAAVGAAGMAVKK147 pKa = 10.35SGMEE151 pKa = 4.22LEE153 pKa = 4.18QQQISMRR160 pKa = 11.84HH161 pKa = 4.12FMGVGNKK168 pKa = 9.42GKK170 pKa = 10.31SSKK173 pKa = 10.53EE174 pKa = 3.68LDD176 pKa = 3.7GMSASYY182 pKa = 11.0LKK184 pKa = 10.53DD185 pKa = 3.17LRR187 pKa = 11.84NNANATPFEE196 pKa = 4.2TGEE199 pKa = 4.29VISAGTRR206 pKa = 11.84SLQIAGGNTKK216 pKa = 10.71DD217 pKa = 3.26AMQMVKK223 pKa = 10.34LAEE226 pKa = 4.7DD227 pKa = 3.64MAALNPGKK235 pKa = 8.56TVGDD239 pKa = 3.67AMEE242 pKa = 5.22ALADD246 pKa = 3.83MNIGEE251 pKa = 4.53MARR254 pKa = 11.84LTEE257 pKa = 4.43FGVKK261 pKa = 10.25ASSTDD266 pKa = 3.47DD267 pKa = 3.52PKK269 pKa = 11.15EE270 pKa = 3.99VQKK273 pKa = 11.17KK274 pKa = 10.2LEE276 pKa = 3.87TMYY279 pKa = 11.28AGGANKK285 pKa = 9.88LAEE288 pKa = 4.68SGSGLLSTIMGKK300 pKa = 10.14LKK302 pKa = 11.12SNIADD307 pKa = 3.2IGLGMLEE314 pKa = 4.03PLKK317 pKa = 10.63PVMTGLIGFIDD328 pKa = 3.69QASPKK333 pKa = 9.45ILEE336 pKa = 4.37VGTKK340 pKa = 7.49ITSGIGMAIGWIQQQMPTLAPIFQTAFGAVSSIVSTVAPIIGQVIGALSPVFMGLLSVASSALSGIASAVKK411 pKa = 8.64TVAPVVSSLISKK423 pKa = 7.97LAPIFLNVGSTLKK436 pKa = 11.14SMGKK440 pKa = 9.07IFKK443 pKa = 10.37NVFDD447 pKa = 4.39SVMKK451 pKa = 10.19IVKK454 pKa = 9.55KK455 pKa = 10.7ASDD458 pKa = 4.07FIKK461 pKa = 10.43PLVNGIIGATKK472 pKa = 10.29GISDD476 pKa = 3.93GVSWVAGKK484 pKa = 9.91LAGNATGTKK493 pKa = 8.26YY494 pKa = 10.08WSGGLSVVGEE504 pKa = 4.49HH505 pKa = 6.58GPEE508 pKa = 3.95LVSMPRR514 pKa = 11.84GSKK517 pKa = 10.23VFTNAEE523 pKa = 4.23SKK525 pKa = 11.46SMINKK530 pKa = 9.38SIPNFRR536 pKa = 11.84QVQGEE541 pKa = 4.12NTNYY545 pKa = 10.71NITIPKK551 pKa = 9.09IAEE554 pKa = 3.91QIVIRR559 pKa = 11.84EE560 pKa = 4.05DD561 pKa = 3.24ADD563 pKa = 3.29IEE565 pKa = 4.78RR566 pKa = 11.84ITSSLIKK573 pKa = 9.61KK574 pKa = 9.18IQMAKK579 pKa = 9.32MGGVVV584 pKa = 2.99
Molecular weight: 62.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
10216 |
44 |
715 |
189.2 |
21.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.08 ± 0.369 | 1.087 ± 0.166 |
5.805 ± 0.268 | 9.397 ± 0.581 |
3.935 ± 0.185 | 5.844 ± 0.552 |
0.989 ± 0.134 | 9.25 ± 0.315 |
10.777 ± 0.498 | 8.232 ± 0.295 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.428 ± 0.233 | 6.725 ± 0.374 |
2.163 ± 0.176 | 2.457 ± 0.187 |
3.534 ± 0.203 | 6.01 ± 0.37 |
5.707 ± 0.463 | 5.403 ± 0.256 |
0.969 ± 0.103 | 4.209 ± 0.351 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
