
Hubei tombus-like virus 17
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.61
Get precalculated fractions of proteins
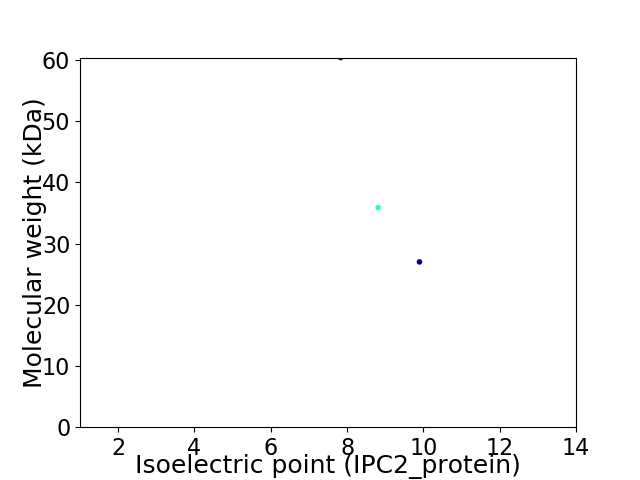
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGE3|A0A1L3KGE3_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 17 OX=1923263 PE=4 SV=1
MM1 pKa = 7.33VSHH4 pKa = 6.95GNQVHH9 pKa = 6.59FNTSEE14 pKa = 4.27CVVIAPSIWTYY25 pKa = 7.26QVRR28 pKa = 11.84VHH30 pKa = 6.56AQCACNQYY38 pKa = 10.04EE39 pKa = 4.45SLIHH43 pKa = 6.27RR44 pKa = 11.84HH45 pKa = 5.34LASKK49 pKa = 10.46EE50 pKa = 3.67VDD52 pKa = 3.3LDD54 pKa = 3.23IGLWRR59 pKa = 11.84RR60 pKa = 11.84LADD63 pKa = 3.3RR64 pKa = 11.84WLHH67 pKa = 6.46IFQADD72 pKa = 3.78LAKK75 pKa = 10.82LSFKK79 pKa = 10.86DD80 pKa = 3.88VISHH84 pKa = 5.78FTGKK88 pKa = 9.9RR89 pKa = 11.84KK90 pKa = 9.66KK91 pKa = 9.77RR92 pKa = 11.84YY93 pKa = 8.96SSAFTHH99 pKa = 5.93YY100 pKa = 10.34CEE102 pKa = 5.32SGYY105 pKa = 9.34QSKK108 pKa = 10.67IKK110 pKa = 10.25MFIKK114 pKa = 9.84QDD116 pKa = 3.54KK117 pKa = 9.57YY118 pKa = 11.19PLDD121 pKa = 4.06TIYY124 pKa = 11.08DD125 pKa = 3.64KK126 pKa = 11.19SPRR129 pKa = 11.84AIQYY133 pKa = 8.55RR134 pKa = 11.84SPEE137 pKa = 3.87FNLAFMQYY145 pKa = 9.8IKK147 pKa = 10.44PIEE150 pKa = 4.12DD151 pKa = 2.73WVYY154 pKa = 11.32NNVTYY159 pKa = 10.71AVVSDD164 pKa = 3.77TPTIAKK170 pKa = 10.06AMNPYY175 pKa = 9.46QRR177 pKa = 11.84AEE179 pKa = 3.56IFRR182 pKa = 11.84YY183 pKa = 8.84KK184 pKa = 10.7VSWFKK189 pKa = 11.12NPKK192 pKa = 9.69FYY194 pKa = 10.87LIDD197 pKa = 3.45HH198 pKa = 6.64SAFDD202 pKa = 3.98SCISKK207 pKa = 10.0YY208 pKa = 10.14HH209 pKa = 6.74LKK211 pKa = 8.01TTHH214 pKa = 6.11KK215 pKa = 10.49KK216 pKa = 9.43YY217 pKa = 10.95FKK219 pKa = 10.75FFDD222 pKa = 3.3NFEE225 pKa = 3.99FRR227 pKa = 11.84KK228 pKa = 10.05LCQAQIHH235 pKa = 6.08NSGKK239 pKa = 7.35TRR241 pKa = 11.84SGIKK245 pKa = 10.29YY246 pKa = 8.88KK247 pKa = 11.05VEE249 pKa = 3.93GTRR252 pKa = 11.84MSGDD256 pKa = 3.54ADD258 pKa = 3.75TALGNCIVNLDD269 pKa = 4.58CITAVLCLSGISKK282 pKa = 10.42YY283 pKa = 11.1DD284 pKa = 3.24IMVDD288 pKa = 3.36GDD290 pKa = 3.86DD291 pKa = 4.37SIVIVEE297 pKa = 3.91QEE299 pKa = 4.0AVIEE303 pKa = 4.1EE304 pKa = 4.28SWFKK308 pKa = 11.14KK309 pKa = 10.43LGFITKK315 pKa = 10.02ISSTTNINEE324 pKa = 4.04VEE326 pKa = 4.44FCQSRR331 pKa = 11.84LCFDD335 pKa = 3.91GSRR338 pKa = 11.84YY339 pKa = 10.56AFVRR343 pKa = 11.84NPMRR347 pKa = 11.84MLAHH351 pKa = 6.43YY352 pKa = 8.19SICNKK357 pKa = 10.01KK358 pKa = 10.71YY359 pKa = 9.6SLKK362 pKa = 10.67QINHH366 pKa = 5.1WLKK369 pKa = 10.63GISMCEE375 pKa = 3.49NSLAGQYY382 pKa = 9.72PIYY385 pKa = 10.36NAIVKK390 pKa = 10.34AFDD393 pKa = 3.1VSDD396 pKa = 3.6RR397 pKa = 11.84FIIDD401 pKa = 3.39EE402 pKa = 4.12EE403 pKa = 4.63LEE405 pKa = 3.85QRR407 pKa = 11.84MKK409 pKa = 10.97GLSFSPLCRR418 pKa = 11.84SITTAARR425 pKa = 11.84KK426 pKa = 9.89SFEE429 pKa = 4.32EE430 pKa = 4.19VWGIDD435 pKa = 3.81HH436 pKa = 6.95ITQEE440 pKa = 4.76LIEE443 pKa = 4.91KK444 pKa = 10.21DD445 pKa = 2.86ILGFKK450 pKa = 10.46GFSKK454 pKa = 11.22VNGKK458 pKa = 9.7YY459 pKa = 10.19DD460 pKa = 3.84EE461 pKa = 5.27PIQRR465 pKa = 11.84AWQRR469 pKa = 11.84YY470 pKa = 4.85QLCHH474 pKa = 6.53EE475 pKa = 4.94SSSSCWRR482 pKa = 11.84DD483 pKa = 3.23CSEE486 pKa = 3.63GRR488 pKa = 11.84YY489 pKa = 10.05GSVGTVVDD497 pKa = 4.32PTDD500 pKa = 4.77DD501 pKa = 4.48PPDD504 pKa = 3.73TTTKK508 pKa = 10.4SLPNGTVGGCPARR521 pKa = 11.84KK522 pKa = 8.93GAAA525 pKa = 3.17
MM1 pKa = 7.33VSHH4 pKa = 6.95GNQVHH9 pKa = 6.59FNTSEE14 pKa = 4.27CVVIAPSIWTYY25 pKa = 7.26QVRR28 pKa = 11.84VHH30 pKa = 6.56AQCACNQYY38 pKa = 10.04EE39 pKa = 4.45SLIHH43 pKa = 6.27RR44 pKa = 11.84HH45 pKa = 5.34LASKK49 pKa = 10.46EE50 pKa = 3.67VDD52 pKa = 3.3LDD54 pKa = 3.23IGLWRR59 pKa = 11.84RR60 pKa = 11.84LADD63 pKa = 3.3RR64 pKa = 11.84WLHH67 pKa = 6.46IFQADD72 pKa = 3.78LAKK75 pKa = 10.82LSFKK79 pKa = 10.86DD80 pKa = 3.88VISHH84 pKa = 5.78FTGKK88 pKa = 9.9RR89 pKa = 11.84KK90 pKa = 9.66KK91 pKa = 9.77RR92 pKa = 11.84YY93 pKa = 8.96SSAFTHH99 pKa = 5.93YY100 pKa = 10.34CEE102 pKa = 5.32SGYY105 pKa = 9.34QSKK108 pKa = 10.67IKK110 pKa = 10.25MFIKK114 pKa = 9.84QDD116 pKa = 3.54KK117 pKa = 9.57YY118 pKa = 11.19PLDD121 pKa = 4.06TIYY124 pKa = 11.08DD125 pKa = 3.64KK126 pKa = 11.19SPRR129 pKa = 11.84AIQYY133 pKa = 8.55RR134 pKa = 11.84SPEE137 pKa = 3.87FNLAFMQYY145 pKa = 9.8IKK147 pKa = 10.44PIEE150 pKa = 4.12DD151 pKa = 2.73WVYY154 pKa = 11.32NNVTYY159 pKa = 10.71AVVSDD164 pKa = 3.77TPTIAKK170 pKa = 10.06AMNPYY175 pKa = 9.46QRR177 pKa = 11.84AEE179 pKa = 3.56IFRR182 pKa = 11.84YY183 pKa = 8.84KK184 pKa = 10.7VSWFKK189 pKa = 11.12NPKK192 pKa = 9.69FYY194 pKa = 10.87LIDD197 pKa = 3.45HH198 pKa = 6.64SAFDD202 pKa = 3.98SCISKK207 pKa = 10.0YY208 pKa = 10.14HH209 pKa = 6.74LKK211 pKa = 8.01TTHH214 pKa = 6.11KK215 pKa = 10.49KK216 pKa = 9.43YY217 pKa = 10.95FKK219 pKa = 10.75FFDD222 pKa = 3.3NFEE225 pKa = 3.99FRR227 pKa = 11.84KK228 pKa = 10.05LCQAQIHH235 pKa = 6.08NSGKK239 pKa = 7.35TRR241 pKa = 11.84SGIKK245 pKa = 10.29YY246 pKa = 8.88KK247 pKa = 11.05VEE249 pKa = 3.93GTRR252 pKa = 11.84MSGDD256 pKa = 3.54ADD258 pKa = 3.75TALGNCIVNLDD269 pKa = 4.58CITAVLCLSGISKK282 pKa = 10.42YY283 pKa = 11.1DD284 pKa = 3.24IMVDD288 pKa = 3.36GDD290 pKa = 3.86DD291 pKa = 4.37SIVIVEE297 pKa = 3.91QEE299 pKa = 4.0AVIEE303 pKa = 4.1EE304 pKa = 4.28SWFKK308 pKa = 11.14KK309 pKa = 10.43LGFITKK315 pKa = 10.02ISSTTNINEE324 pKa = 4.04VEE326 pKa = 4.44FCQSRR331 pKa = 11.84LCFDD335 pKa = 3.91GSRR338 pKa = 11.84YY339 pKa = 10.56AFVRR343 pKa = 11.84NPMRR347 pKa = 11.84MLAHH351 pKa = 6.43YY352 pKa = 8.19SICNKK357 pKa = 10.01KK358 pKa = 10.71YY359 pKa = 9.6SLKK362 pKa = 10.67QINHH366 pKa = 5.1WLKK369 pKa = 10.63GISMCEE375 pKa = 3.49NSLAGQYY382 pKa = 9.72PIYY385 pKa = 10.36NAIVKK390 pKa = 10.34AFDD393 pKa = 3.1VSDD396 pKa = 3.6RR397 pKa = 11.84FIIDD401 pKa = 3.39EE402 pKa = 4.12EE403 pKa = 4.63LEE405 pKa = 3.85QRR407 pKa = 11.84MKK409 pKa = 10.97GLSFSPLCRR418 pKa = 11.84SITTAARR425 pKa = 11.84KK426 pKa = 9.89SFEE429 pKa = 4.32EE430 pKa = 4.19VWGIDD435 pKa = 3.81HH436 pKa = 6.95ITQEE440 pKa = 4.76LIEE443 pKa = 4.91KK444 pKa = 10.21DD445 pKa = 2.86ILGFKK450 pKa = 10.46GFSKK454 pKa = 11.22VNGKK458 pKa = 9.7YY459 pKa = 10.19DD460 pKa = 3.84EE461 pKa = 5.27PIQRR465 pKa = 11.84AWQRR469 pKa = 11.84YY470 pKa = 4.85QLCHH474 pKa = 6.53EE475 pKa = 4.94SSSSCWRR482 pKa = 11.84DD483 pKa = 3.23CSEE486 pKa = 3.63GRR488 pKa = 11.84YY489 pKa = 10.05GSVGTVVDD497 pKa = 4.32PTDD500 pKa = 4.77DD501 pKa = 4.48PPDD504 pKa = 3.73TTTKK508 pKa = 10.4SLPNGTVGGCPARR521 pKa = 11.84KK522 pKa = 8.93GAAA525 pKa = 3.17
Molecular weight: 60.37 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFY9|A0A1L3KFY9_9VIRU RNA-directed RNA polymerase OS=Hubei tombus-like virus 17 OX=1923263 PE=4 SV=1
MM1 pKa = 7.05MNQYY5 pKa = 10.18RR6 pKa = 11.84GRR8 pKa = 11.84GRR10 pKa = 11.84GTNFAMNLAAAAGGTAARR28 pKa = 11.84VAMDD32 pKa = 5.32RR33 pKa = 11.84LGQWWTQPTTPPTPQPNPYY52 pKa = 10.8QMGQLVAALPGRR64 pKa = 11.84GRR66 pKa = 11.84PRR68 pKa = 11.84RR69 pKa = 11.84GRR71 pKa = 11.84GRR73 pKa = 11.84GRR75 pKa = 11.84GRR77 pKa = 11.84GRR79 pKa = 11.84RR80 pKa = 11.84PQVARR85 pKa = 11.84SGGQPSSGVQTRR97 pKa = 11.84SGDD100 pKa = 3.6LFVARR105 pKa = 11.84GTEE108 pKa = 3.82VLGPVTGEE116 pKa = 3.91LQVLEE121 pKa = 5.06FNPSCDD127 pKa = 3.32GLPRR131 pKa = 11.84LAAVEE136 pKa = 4.14KK137 pKa = 9.17MYY139 pKa = 10.66HH140 pKa = 6.62RR141 pKa = 11.84YY142 pKa = 9.31RR143 pKa = 11.84IKK145 pKa = 10.58YY146 pKa = 8.83VNIAFKK152 pKa = 10.61SGSGTATAGNVAFGVCVGPKK172 pKa = 8.59VANVKK177 pKa = 7.87TQSDD181 pKa = 3.52IMKK184 pKa = 9.55LRR186 pKa = 11.84PFSYY190 pKa = 10.44VPAWKK195 pKa = 10.23NSSITVGSDD204 pKa = 2.39IDD206 pKa = 3.14IGRR209 pKa = 11.84FMISGSKK216 pKa = 10.39SEE218 pKa = 4.86DD219 pKa = 2.99GVAFTLYY226 pKa = 9.85IFASAAQLGVIQISYY241 pKa = 9.62EE242 pKa = 4.13VEE244 pKa = 4.35FSHH247 pKa = 6.51PTPFF251 pKa = 4.63
MM1 pKa = 7.05MNQYY5 pKa = 10.18RR6 pKa = 11.84GRR8 pKa = 11.84GRR10 pKa = 11.84GTNFAMNLAAAAGGTAARR28 pKa = 11.84VAMDD32 pKa = 5.32RR33 pKa = 11.84LGQWWTQPTTPPTPQPNPYY52 pKa = 10.8QMGQLVAALPGRR64 pKa = 11.84GRR66 pKa = 11.84PRR68 pKa = 11.84RR69 pKa = 11.84GRR71 pKa = 11.84GRR73 pKa = 11.84GRR75 pKa = 11.84GRR77 pKa = 11.84GRR79 pKa = 11.84RR80 pKa = 11.84PQVARR85 pKa = 11.84SGGQPSSGVQTRR97 pKa = 11.84SGDD100 pKa = 3.6LFVARR105 pKa = 11.84GTEE108 pKa = 3.82VLGPVTGEE116 pKa = 3.91LQVLEE121 pKa = 5.06FNPSCDD127 pKa = 3.32GLPRR131 pKa = 11.84LAAVEE136 pKa = 4.14KK137 pKa = 9.17MYY139 pKa = 10.66HH140 pKa = 6.62RR141 pKa = 11.84YY142 pKa = 9.31RR143 pKa = 11.84IKK145 pKa = 10.58YY146 pKa = 8.83VNIAFKK152 pKa = 10.61SGSGTATAGNVAFGVCVGPKK172 pKa = 8.59VANVKK177 pKa = 7.87TQSDD181 pKa = 3.52IMKK184 pKa = 9.55LRR186 pKa = 11.84PFSYY190 pKa = 10.44VPAWKK195 pKa = 10.23NSSITVGSDD204 pKa = 2.39IDD206 pKa = 3.14IGRR209 pKa = 11.84FMISGSKK216 pKa = 10.39SEE218 pKa = 4.86DD219 pKa = 2.99GVAFTLYY226 pKa = 9.85IFASAAQLGVIQISYY241 pKa = 9.62EE242 pKa = 4.13VEE244 pKa = 4.35FSHH247 pKa = 6.51PTPFF251 pKa = 4.63
Molecular weight: 27.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1078 |
251 |
525 |
359.3 |
41.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.308 ± 1.035 | 2.505 ± 0.646 |
4.36 ± 1.033 | 5.009 ± 0.776 |
4.638 ± 0.424 | 6.215 ± 2.054 |
3.061 ± 0.779 | 6.215 ± 1.007 |
5.937 ± 1.287 | 7.05 ± 1.271 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.69 ± 0.468 | 4.267 ± 0.343 |
5.38 ± 1.273 | 4.917 ± 0.565 |
7.143 ± 1.199 | 7.05 ± 1.13 |
5.195 ± 0.249 | 6.308 ± 0.542 |
1.763 ± 0.178 | 3.989 ± 0.572 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
