
Musca hytrovirus(isolate Musca domestica/United States/Boucias/-) (MHV)
Taxonomy: Viruses; Naldaviricetes; Lefavirales; Hytrosaviridae; Muscavirus; Musca hytrosavirus
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins
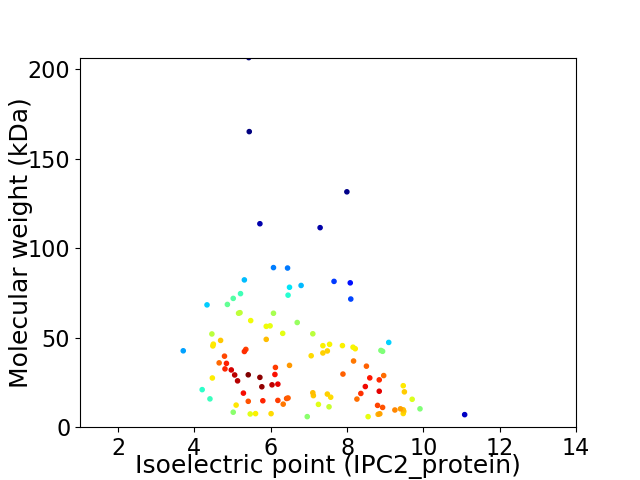
Virtual 2D-PAGE plot for 108 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B2YG26|B2YG26_MHVB Ac150-like protein OS=Musca hytrovirus(isolate Musca domestica/United States/Boucias/-) OX=523909 GN=MdSGHV049 PE=4 SV=1
MM1 pKa = 7.3SRR3 pKa = 11.84NFAFFLFLGICLVASRR19 pKa = 11.84VVDD22 pKa = 3.95CQEE25 pKa = 3.74GTEE28 pKa = 4.03DD29 pKa = 3.27QTVNEE34 pKa = 4.42TQGDD38 pKa = 3.66QSVNEE43 pKa = 4.14NPGDD47 pKa = 3.71QSVDD51 pKa = 3.27EE52 pKa = 4.52AQGDD56 pKa = 3.64QSVNEE61 pKa = 4.14NPGDD65 pKa = 3.59QSVYY69 pKa = 9.96EE70 pKa = 4.13AQGDD74 pKa = 3.72QSVNEE79 pKa = 4.19VPEE82 pKa = 4.4DD83 pKa = 3.71QPADD87 pKa = 3.79DD88 pKa = 4.5VADD91 pKa = 4.48PVPDD95 pKa = 4.43DD96 pKa = 4.13QPADD100 pKa = 3.82DD101 pKa = 4.83VGSEE105 pKa = 4.39STNSPHH111 pKa = 7.66EE112 pKa = 3.86ITEE115 pKa = 4.22PDD117 pKa = 3.23ASGDD121 pKa = 3.76GQASEE126 pKa = 4.68DD127 pKa = 3.5QSAPTEE133 pKa = 4.2APGDD137 pKa = 3.52QSAPTEE143 pKa = 4.28APGDD147 pKa = 3.88QPVDD151 pKa = 3.64EE152 pKa = 5.28APGASEE158 pKa = 3.97NEE160 pKa = 4.1KK161 pKa = 10.72KK162 pKa = 10.71EE163 pKa = 4.12EE164 pKa = 4.05EE165 pKa = 4.19EE166 pKa = 4.28EE167 pKa = 4.17PAVVPDD173 pKa = 4.03GDD175 pKa = 4.23GSGDD179 pKa = 3.52VNNDD183 pKa = 3.38SNHH186 pKa = 6.07SNSTEE191 pKa = 3.95VAVEE195 pKa = 4.2DD196 pKa = 4.62NSDD199 pKa = 3.63HH200 pKa = 8.12VDD202 pKa = 3.59DD203 pKa = 4.86QSEE206 pKa = 4.31NNPGGRR212 pKa = 11.84DD213 pKa = 3.62EE214 pKa = 5.31VPTEE218 pKa = 4.28SPPEE222 pKa = 4.11DD223 pKa = 3.06TSAPEE228 pKa = 4.15NIEE231 pKa = 4.12DD232 pKa = 4.27EE233 pKa = 4.27KK234 pKa = 11.47SKK236 pKa = 11.25YY237 pKa = 10.65LGDD240 pKa = 3.6EE241 pKa = 4.23YY242 pKa = 11.36NFFMEE247 pKa = 4.55HH248 pKa = 6.83CSPSEE253 pKa = 4.19SVNHH257 pKa = 6.66FGYY260 pKa = 10.8EE261 pKa = 3.72LLYY264 pKa = 11.1DD265 pKa = 3.64FFINVLHH272 pKa = 6.92IFYY275 pKa = 10.24SDD277 pKa = 3.51HH278 pKa = 7.38ALDD281 pKa = 5.04GYY283 pKa = 11.12NSNRR287 pKa = 11.84TIQKK291 pKa = 8.74YY292 pKa = 9.81FGRR295 pKa = 11.84DD296 pKa = 2.9VYY298 pKa = 11.5DD299 pKa = 3.42QVFNDD304 pKa = 3.57YY305 pKa = 11.53ANAHH309 pKa = 6.03SLSEE313 pKa = 4.11PLMSCPEE320 pKa = 3.87NVEE323 pKa = 4.26EE324 pKa = 5.13KK325 pKa = 10.34FKK327 pKa = 11.33KK328 pKa = 10.33SINEE332 pKa = 4.45FIVHH336 pKa = 6.95ALSTLLTIYY345 pKa = 10.93YY346 pKa = 9.2YY347 pKa = 10.45GTVSADD353 pKa = 3.45DD354 pKa = 3.41IQRR357 pKa = 11.84IEE359 pKa = 4.14EE360 pKa = 4.23TLSAGQQIDD369 pKa = 3.89SLIDD373 pKa = 3.63GYY375 pKa = 11.62GQMSGVLMDD384 pKa = 4.49LCRR387 pKa = 11.84KK388 pKa = 9.17CHH390 pKa = 5.73
MM1 pKa = 7.3SRR3 pKa = 11.84NFAFFLFLGICLVASRR19 pKa = 11.84VVDD22 pKa = 3.95CQEE25 pKa = 3.74GTEE28 pKa = 4.03DD29 pKa = 3.27QTVNEE34 pKa = 4.42TQGDD38 pKa = 3.66QSVNEE43 pKa = 4.14NPGDD47 pKa = 3.71QSVDD51 pKa = 3.27EE52 pKa = 4.52AQGDD56 pKa = 3.64QSVNEE61 pKa = 4.14NPGDD65 pKa = 3.59QSVYY69 pKa = 9.96EE70 pKa = 4.13AQGDD74 pKa = 3.72QSVNEE79 pKa = 4.19VPEE82 pKa = 4.4DD83 pKa = 3.71QPADD87 pKa = 3.79DD88 pKa = 4.5VADD91 pKa = 4.48PVPDD95 pKa = 4.43DD96 pKa = 4.13QPADD100 pKa = 3.82DD101 pKa = 4.83VGSEE105 pKa = 4.39STNSPHH111 pKa = 7.66EE112 pKa = 3.86ITEE115 pKa = 4.22PDD117 pKa = 3.23ASGDD121 pKa = 3.76GQASEE126 pKa = 4.68DD127 pKa = 3.5QSAPTEE133 pKa = 4.2APGDD137 pKa = 3.52QSAPTEE143 pKa = 4.28APGDD147 pKa = 3.88QPVDD151 pKa = 3.64EE152 pKa = 5.28APGASEE158 pKa = 3.97NEE160 pKa = 4.1KK161 pKa = 10.72KK162 pKa = 10.71EE163 pKa = 4.12EE164 pKa = 4.05EE165 pKa = 4.19EE166 pKa = 4.28EE167 pKa = 4.17PAVVPDD173 pKa = 4.03GDD175 pKa = 4.23GSGDD179 pKa = 3.52VNNDD183 pKa = 3.38SNHH186 pKa = 6.07SNSTEE191 pKa = 3.95VAVEE195 pKa = 4.2DD196 pKa = 4.62NSDD199 pKa = 3.63HH200 pKa = 8.12VDD202 pKa = 3.59DD203 pKa = 4.86QSEE206 pKa = 4.31NNPGGRR212 pKa = 11.84DD213 pKa = 3.62EE214 pKa = 5.31VPTEE218 pKa = 4.28SPPEE222 pKa = 4.11DD223 pKa = 3.06TSAPEE228 pKa = 4.15NIEE231 pKa = 4.12DD232 pKa = 4.27EE233 pKa = 4.27KK234 pKa = 11.47SKK236 pKa = 11.25YY237 pKa = 10.65LGDD240 pKa = 3.6EE241 pKa = 4.23YY242 pKa = 11.36NFFMEE247 pKa = 4.55HH248 pKa = 6.83CSPSEE253 pKa = 4.19SVNHH257 pKa = 6.66FGYY260 pKa = 10.8EE261 pKa = 3.72LLYY264 pKa = 11.1DD265 pKa = 3.64FFINVLHH272 pKa = 6.92IFYY275 pKa = 10.24SDD277 pKa = 3.51HH278 pKa = 7.38ALDD281 pKa = 5.04GYY283 pKa = 11.12NSNRR287 pKa = 11.84TIQKK291 pKa = 8.74YY292 pKa = 9.81FGRR295 pKa = 11.84DD296 pKa = 2.9VYY298 pKa = 11.5DD299 pKa = 3.42QVFNDD304 pKa = 3.57YY305 pKa = 11.53ANAHH309 pKa = 6.03SLSEE313 pKa = 4.11PLMSCPEE320 pKa = 3.87NVEE323 pKa = 4.26EE324 pKa = 5.13KK325 pKa = 10.34FKK327 pKa = 11.33KK328 pKa = 10.33SINEE332 pKa = 4.45FIVHH336 pKa = 6.95ALSTLLTIYY345 pKa = 10.93YY346 pKa = 9.2YY347 pKa = 10.45GTVSADD353 pKa = 3.45DD354 pKa = 3.41IQRR357 pKa = 11.84IEE359 pKa = 4.14EE360 pKa = 4.23TLSAGQQIDD369 pKa = 3.89SLIDD373 pKa = 3.63GYY375 pKa = 11.62GQMSGVLMDD384 pKa = 4.49LCRR387 pKa = 11.84KK388 pKa = 9.17CHH390 pKa = 5.73
Molecular weight: 42.67 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B2YG04|B2YG04_MHVB PDDEXK_1 domain-containing protein OS=Musca hytrovirus(isolate Musca domestica/United States/Boucias/-) OX=523909 GN=MdSGHV027 PE=4 SV=1
MM1 pKa = 7.7AKK3 pKa = 10.44ASLDD7 pKa = 3.81SISRR11 pKa = 11.84NMRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84KK17 pKa = 9.61RR18 pKa = 11.84KK19 pKa = 8.97SSSSSSSSSSSSSKK33 pKa = 10.11RR34 pKa = 11.84RR35 pKa = 11.84TPRR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 9.43SSSKK44 pKa = 10.2RR45 pKa = 11.84RR46 pKa = 11.84TTTRR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84APAYY58 pKa = 10.13RR59 pKa = 11.84RR60 pKa = 3.57
MM1 pKa = 7.7AKK3 pKa = 10.44ASLDD7 pKa = 3.81SISRR11 pKa = 11.84NMRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84KK17 pKa = 9.61RR18 pKa = 11.84KK19 pKa = 8.97SSSSSSSSSSSSSKK33 pKa = 10.11RR34 pKa = 11.84RR35 pKa = 11.84TPRR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 9.43SSSKK44 pKa = 10.2RR45 pKa = 11.84RR46 pKa = 11.84TTTRR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84APAYY58 pKa = 10.13RR59 pKa = 11.84RR60 pKa = 3.57
Molecular weight: 7.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
37570 |
51 |
1780 |
347.9 |
39.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.961 ± 0.147 | 2.036 ± 0.142 |
6.42 ± 0.207 | 5.193 ± 0.162 |
4.847 ± 0.138 | 4.357 ± 0.201 |
2.31 ± 0.125 | 6.939 ± 0.154 |
4.884 ± 0.184 | 9.084 ± 0.233 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.05 ± 0.107 | 6.537 ± 0.207 |
4.064 ± 0.167 | 3.7 ± 0.111 |
6.114 ± 0.208 | 7.49 ± 0.217 |
6.268 ± 0.188 | 6.22 ± 0.142 |
0.735 ± 0.053 | 4.788 ± 0.162 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
