
Marinobacter sp. Z-D5-3
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Alteromonadaceae; Marinobacter; unclassified Marinobacter
Average proteome isoelectric point is 5.88
Get precalculated fractions of proteins
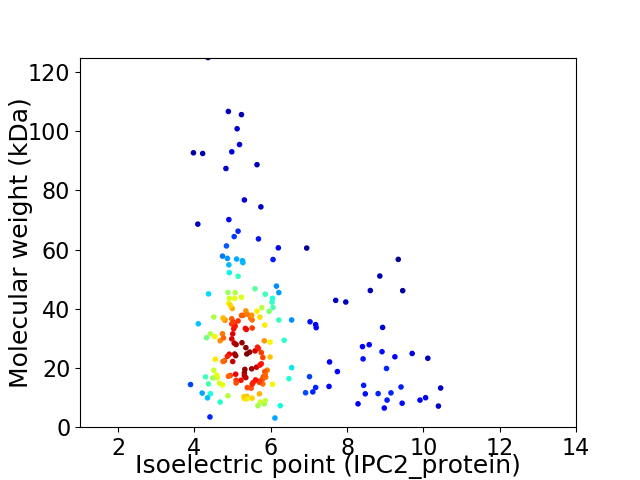
Virtual 2D-PAGE plot for 201 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2T4CLF5|A0A2T4CLF5_9ALTE Chaperone protein ClpB OS=Marinobacter sp. Z-D5-3 OX=2137200 GN=clpB PE=3 SV=1
MM1 pKa = 7.37FKK3 pKa = 9.48KK4 pKa = 9.12TLISLAVASSVGLTGCFDD22 pKa = 3.72SGEE25 pKa = 4.29TGANANPQYY34 pKa = 10.96NIVDD38 pKa = 3.77TTIDD42 pKa = 3.34RR43 pKa = 11.84SIVRR47 pKa = 11.84PVFDD51 pKa = 5.77PNPISPTVGFPANFDD66 pKa = 4.02LLLLLGATQSSNYY79 pKa = 10.03DD80 pKa = 3.35FTGLTSGTDD89 pKa = 3.5PASNAINDD97 pKa = 4.01LAGFSTSGQFNIAFDD112 pKa = 4.1GSLNPDD118 pKa = 3.66TVIKK122 pKa = 9.97GQTVFLVPLNVAPAVDD138 pKa = 4.38SAPLALPSVNPASVTGVDD156 pKa = 3.77TTPEE160 pKa = 3.83NQPNYY165 pKa = 8.51RR166 pKa = 11.84VEE168 pKa = 5.08VITQDD173 pKa = 2.98GVTNNTIRR181 pKa = 11.84ITPLEE186 pKa = 4.26PLLEE190 pKa = 4.29SKK192 pKa = 10.47KK193 pKa = 10.68YY194 pKa = 10.19IVIVSDD200 pKa = 3.79GVTGANGKK208 pKa = 7.82PTEE211 pKa = 4.03QSIAAQQLTTGPLGSEE227 pKa = 3.86NLSNIRR233 pKa = 11.84DD234 pKa = 3.75LVNALNGLGQQIISATGKK252 pKa = 10.21DD253 pKa = 3.17VALAYY258 pKa = 9.05TFTTAGQSTVLKK270 pKa = 10.82NMASPGTYY278 pKa = 9.91LQALGQKK285 pKa = 9.61IGFTALLKK293 pKa = 10.51AVRR296 pKa = 11.84DD297 pKa = 3.98NNPNANFSQLTEE309 pKa = 3.96ILGEE313 pKa = 4.0IQDD316 pKa = 4.57GNVEE320 pKa = 4.76DD321 pKa = 4.5YY322 pKa = 10.85PEE324 pKa = 3.85TAAAVQEE331 pKa = 4.26AASVATSTNIQNAIGASFSTGALHH355 pKa = 7.45FPTTRR360 pKa = 11.84PSFFYY365 pKa = 11.06SKK367 pKa = 10.43DD368 pKa = 3.35APATGLATIANLPDD382 pKa = 3.3GNAIKK387 pKa = 10.49SAAAGVSVSQGSIVLPYY404 pKa = 8.53YY405 pKa = 10.33QPIPAVSGGDD415 pKa = 3.46ALVSRR420 pKa = 11.84GWQGDD425 pKa = 3.95TNLEE429 pKa = 3.92TSLNSNLPASGTTFQFLRR447 pKa = 11.84DD448 pKa = 3.12IDD450 pKa = 3.6GTLNVNSNFPYY461 pKa = 10.28PKK463 pKa = 10.09VEE465 pKa = 4.16GNVAVPVVAFYY476 pKa = 10.67PNAGICGGGTIAGVTILQHH495 pKa = 6.75GITVDD500 pKa = 3.54RR501 pKa = 11.84SVSMLPGILIAASTCQAVIAIDD523 pKa = 3.78QPLHH527 pKa = 5.78GLAGATAGTLPGLDD541 pKa = 3.3VLTAEE546 pKa = 4.81DD547 pKa = 3.71VGTAFDD553 pKa = 4.29SVPGLAYY560 pKa = 9.62IGEE563 pKa = 4.18RR564 pKa = 11.84HH565 pKa = 6.13FNYY568 pKa = 10.06TDD570 pKa = 3.56AGSLTPVQAASIADD584 pKa = 3.73VEE586 pKa = 4.68SGSLYY591 pKa = 11.26VNLKK595 pKa = 9.57SLQGARR601 pKa = 11.84DD602 pKa = 3.72NNRR605 pKa = 11.84QGVLDD610 pKa = 4.72LLNLSASVDD619 pKa = 3.6GSTSPQGISGLDD631 pKa = 3.1IRR633 pKa = 11.84GDD635 pKa = 3.71ANPDD639 pKa = 3.41FTSTTPVSFIGHH651 pKa = 5.68SLGGITGTTFASLSNDD667 pKa = 3.31PAVRR671 pKa = 11.84GAYY674 pKa = 8.27AAPGLPPIQFPEE686 pKa = 4.2LEE688 pKa = 4.22NVSLHH693 pKa = 5.1NTGGQIAKK701 pKa = 10.03LVEE704 pKa = 4.07NSQSISSQVLPALPQQGTSDD724 pKa = 4.5LEE726 pKa = 4.06TFLYY730 pKa = 10.52VFQSVLDD737 pKa = 3.85EE738 pKa = 4.33VDD740 pKa = 3.06PVTYY744 pKa = 10.29AKK746 pKa = 10.54SLGASEE752 pKa = 4.52TNLLVTEE759 pKa = 4.63VIGDD763 pKa = 3.6STVPNEE769 pKa = 4.35ANVNPLGSALSAPLAGTEE787 pKa = 3.87PLMALFDD794 pKa = 4.46LGAGGTNLADD804 pKa = 3.47GTALNVVDD812 pKa = 5.43SNTAPTADD820 pKa = 3.82AGLPAAVFFAGSNPCAQANHH840 pKa = 5.63GTFVVPQAPNDD851 pKa = 3.59QCPGGIANTIDD862 pKa = 3.51AFSAMVTQTAQAIAMEE878 pKa = 4.27QLPVDD883 pKa = 3.88AAIQATLGQSPTIDD897 pKa = 3.24STLDD901 pKa = 3.13QDD903 pKa = 3.79QQ904 pKa = 3.43
MM1 pKa = 7.37FKK3 pKa = 9.48KK4 pKa = 9.12TLISLAVASSVGLTGCFDD22 pKa = 3.72SGEE25 pKa = 4.29TGANANPQYY34 pKa = 10.96NIVDD38 pKa = 3.77TTIDD42 pKa = 3.34RR43 pKa = 11.84SIVRR47 pKa = 11.84PVFDD51 pKa = 5.77PNPISPTVGFPANFDD66 pKa = 4.02LLLLLGATQSSNYY79 pKa = 10.03DD80 pKa = 3.35FTGLTSGTDD89 pKa = 3.5PASNAINDD97 pKa = 4.01LAGFSTSGQFNIAFDD112 pKa = 4.1GSLNPDD118 pKa = 3.66TVIKK122 pKa = 9.97GQTVFLVPLNVAPAVDD138 pKa = 4.38SAPLALPSVNPASVTGVDD156 pKa = 3.77TTPEE160 pKa = 3.83NQPNYY165 pKa = 8.51RR166 pKa = 11.84VEE168 pKa = 5.08VITQDD173 pKa = 2.98GVTNNTIRR181 pKa = 11.84ITPLEE186 pKa = 4.26PLLEE190 pKa = 4.29SKK192 pKa = 10.47KK193 pKa = 10.68YY194 pKa = 10.19IVIVSDD200 pKa = 3.79GVTGANGKK208 pKa = 7.82PTEE211 pKa = 4.03QSIAAQQLTTGPLGSEE227 pKa = 3.86NLSNIRR233 pKa = 11.84DD234 pKa = 3.75LVNALNGLGQQIISATGKK252 pKa = 10.21DD253 pKa = 3.17VALAYY258 pKa = 9.05TFTTAGQSTVLKK270 pKa = 10.82NMASPGTYY278 pKa = 9.91LQALGQKK285 pKa = 9.61IGFTALLKK293 pKa = 10.51AVRR296 pKa = 11.84DD297 pKa = 3.98NNPNANFSQLTEE309 pKa = 3.96ILGEE313 pKa = 4.0IQDD316 pKa = 4.57GNVEE320 pKa = 4.76DD321 pKa = 4.5YY322 pKa = 10.85PEE324 pKa = 3.85TAAAVQEE331 pKa = 4.26AASVATSTNIQNAIGASFSTGALHH355 pKa = 7.45FPTTRR360 pKa = 11.84PSFFYY365 pKa = 11.06SKK367 pKa = 10.43DD368 pKa = 3.35APATGLATIANLPDD382 pKa = 3.3GNAIKK387 pKa = 10.49SAAAGVSVSQGSIVLPYY404 pKa = 8.53YY405 pKa = 10.33QPIPAVSGGDD415 pKa = 3.46ALVSRR420 pKa = 11.84GWQGDD425 pKa = 3.95TNLEE429 pKa = 3.92TSLNSNLPASGTTFQFLRR447 pKa = 11.84DD448 pKa = 3.12IDD450 pKa = 3.6GTLNVNSNFPYY461 pKa = 10.28PKK463 pKa = 10.09VEE465 pKa = 4.16GNVAVPVVAFYY476 pKa = 10.67PNAGICGGGTIAGVTILQHH495 pKa = 6.75GITVDD500 pKa = 3.54RR501 pKa = 11.84SVSMLPGILIAASTCQAVIAIDD523 pKa = 3.78QPLHH527 pKa = 5.78GLAGATAGTLPGLDD541 pKa = 3.3VLTAEE546 pKa = 4.81DD547 pKa = 3.71VGTAFDD553 pKa = 4.29SVPGLAYY560 pKa = 9.62IGEE563 pKa = 4.18RR564 pKa = 11.84HH565 pKa = 6.13FNYY568 pKa = 10.06TDD570 pKa = 3.56AGSLTPVQAASIADD584 pKa = 3.73VEE586 pKa = 4.68SGSLYY591 pKa = 11.26VNLKK595 pKa = 9.57SLQGARR601 pKa = 11.84DD602 pKa = 3.72NNRR605 pKa = 11.84QGVLDD610 pKa = 4.72LLNLSASVDD619 pKa = 3.6GSTSPQGISGLDD631 pKa = 3.1IRR633 pKa = 11.84GDD635 pKa = 3.71ANPDD639 pKa = 3.41FTSTTPVSFIGHH651 pKa = 5.68SLGGITGTTFASLSNDD667 pKa = 3.31PAVRR671 pKa = 11.84GAYY674 pKa = 8.27AAPGLPPIQFPEE686 pKa = 4.2LEE688 pKa = 4.22NVSLHH693 pKa = 5.1NTGGQIAKK701 pKa = 10.03LVEE704 pKa = 4.07NSQSISSQVLPALPQQGTSDD724 pKa = 4.5LEE726 pKa = 4.06TFLYY730 pKa = 10.52VFQSVLDD737 pKa = 3.85EE738 pKa = 4.33VDD740 pKa = 3.06PVTYY744 pKa = 10.29AKK746 pKa = 10.54SLGASEE752 pKa = 4.52TNLLVTEE759 pKa = 4.63VIGDD763 pKa = 3.6STVPNEE769 pKa = 4.35ANVNPLGSALSAPLAGTEE787 pKa = 3.87PLMALFDD794 pKa = 4.46LGAGGTNLADD804 pKa = 3.47GTALNVVDD812 pKa = 5.43SNTAPTADD820 pKa = 3.82AGLPAAVFFAGSNPCAQANHH840 pKa = 5.63GTFVVPQAPNDD851 pKa = 3.59QCPGGIANTIDD862 pKa = 3.51AFSAMVTQTAQAIAMEE878 pKa = 4.27QLPVDD883 pKa = 3.88AAIQATLGQSPTIDD897 pKa = 3.24STLDD901 pKa = 3.13QDD903 pKa = 3.79QQ904 pKa = 3.43
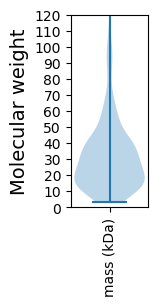
Molecular weight: 92.67 kDa
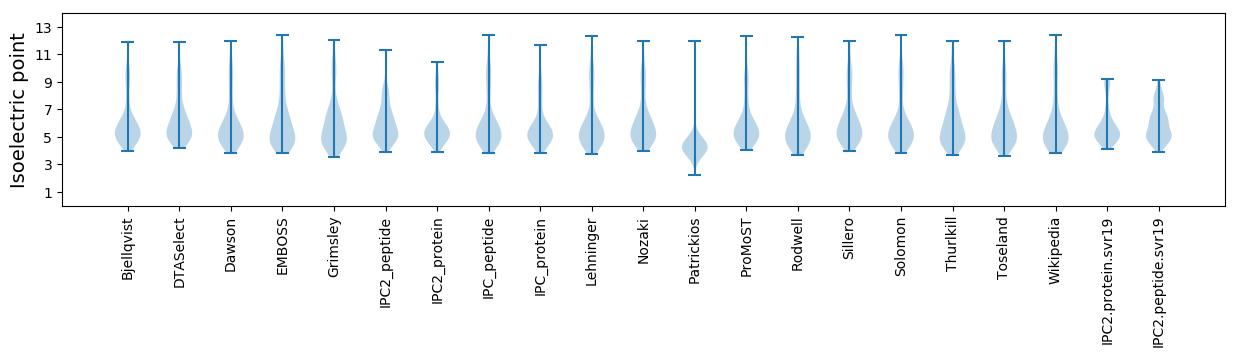
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2T4CLE1|A0A2T4CLE1_9ALTE Crp/Fnr family transcriptional regulator OS=Marinobacter sp. Z-D5-3 OX=2137200 GN=C9984_00050 PE=4 SV=1
MM1 pKa = 7.57ANSPQAKK8 pKa = 9.2KK9 pKa = 10.21RR10 pKa = 11.84ARR12 pKa = 11.84QNEE15 pKa = 4.12KK16 pKa = 10.14NRR18 pKa = 11.84KK19 pKa = 8.17HH20 pKa = 5.45NASLRR25 pKa = 11.84SMARR29 pKa = 11.84TYY31 pKa = 10.76VKK33 pKa = 10.39KK34 pKa = 10.26IQSKK38 pKa = 10.01IEE40 pKa = 3.9AGNYY44 pKa = 9.62EE45 pKa = 4.01EE46 pKa = 5.09AQAAFQQAQPILDD59 pKa = 3.6SMVNKK64 pKa = 10.8GIFAKK69 pKa = 10.82NKK71 pKa = 7.26VARR74 pKa = 11.84SKK76 pKa = 11.12SRR78 pKa = 11.84LSAKK82 pKa = 9.95IKK84 pKa = 10.18ALKK87 pKa = 9.95SAA89 pKa = 4.08
MM1 pKa = 7.57ANSPQAKK8 pKa = 9.2KK9 pKa = 10.21RR10 pKa = 11.84ARR12 pKa = 11.84QNEE15 pKa = 4.12KK16 pKa = 10.14NRR18 pKa = 11.84KK19 pKa = 8.17HH20 pKa = 5.45NASLRR25 pKa = 11.84SMARR29 pKa = 11.84TYY31 pKa = 10.76VKK33 pKa = 10.39KK34 pKa = 10.26IQSKK38 pKa = 10.01IEE40 pKa = 3.9AGNYY44 pKa = 9.62EE45 pKa = 4.01EE46 pKa = 5.09AQAAFQQAQPILDD59 pKa = 3.6SMVNKK64 pKa = 10.8GIFAKK69 pKa = 10.82NKK71 pKa = 7.26VARR74 pKa = 11.84SKK76 pKa = 11.12SRR78 pKa = 11.84LSAKK82 pKa = 9.95IKK84 pKa = 10.18ALKK87 pKa = 9.95SAA89 pKa = 4.08
Molecular weight: 9.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
58420 |
28 |
1159 |
290.6 |
31.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.949 ± 0.186 | 0.828 ± 0.045 |
5.565 ± 0.121 | 6.768 ± 0.193 |
3.826 ± 0.12 | 8.025 ± 0.168 |
1.953 ± 0.089 | 5.404 ± 0.12 |
3.566 ± 0.155 | 10.765 ± 0.241 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.491 ± 0.105 | 3.487 ± 0.148 |
4.575 ± 0.112 | 4.06 ± 0.139 |
6.301 ± 0.161 | 5.847 ± 0.175 |
5.262 ± 0.139 | 7.432 ± 0.14 |
1.392 ± 0.075 | 2.504 ± 0.104 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
