
Halopiger salifodinae
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Natrialbales; Natrialbaceae; Halopiger
Average proteome isoelectric point is 4.8
Get precalculated fractions of proteins
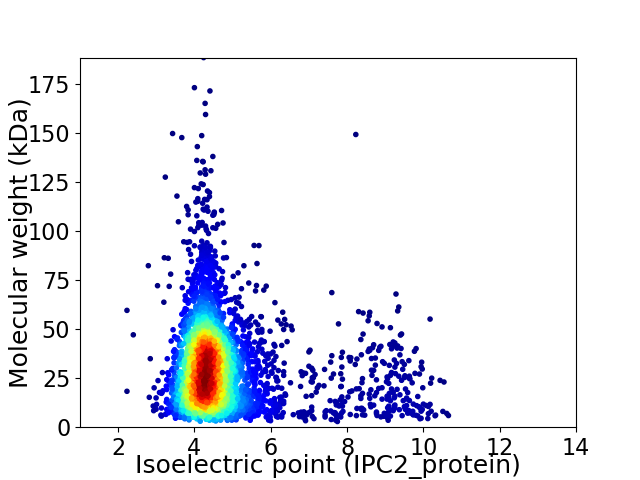
Virtual 2D-PAGE plot for 4090 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I0NPH5|A0A1I0NPH5_9EURY Kynurenine formamidase OS=Halopiger salifodinae OX=1202768 GN=SAMN05216285_1923 PE=4 SV=1
MM1 pKa = 7.7ANDD4 pKa = 4.68DD5 pKa = 4.12NNTSKK10 pKa = 9.2TCRR13 pKa = 11.84IGKK16 pKa = 7.86TSVGRR21 pKa = 11.84RR22 pKa = 11.84TFLGAAGAGAVATTLAGCLGGGDD45 pKa = 4.52DD46 pKa = 5.23DD47 pKa = 5.43GFTIGHH53 pKa = 7.6LAPLEE58 pKa = 4.02NTQGLGSEE66 pKa = 4.47QSAEE70 pKa = 3.95LAAKK74 pKa = 9.33EE75 pKa = 4.15INDD78 pKa = 3.93DD79 pKa = 3.6GGIRR83 pKa = 11.84DD84 pKa = 3.93EE85 pKa = 4.41DD86 pKa = 4.12VEE88 pKa = 4.43IVSADD93 pKa = 3.48TRR95 pKa = 11.84SDD97 pKa = 3.2PSTAQDD103 pKa = 3.29EE104 pKa = 4.46ASRR107 pKa = 11.84LINQEE112 pKa = 3.82RR113 pKa = 11.84VDD115 pKa = 4.12LLVGTFSSEE124 pKa = 3.85VSVNIMDD131 pKa = 5.01MIAEE135 pKa = 4.07NDD137 pKa = 3.17VPYY140 pKa = 10.31IVTGSASPAIIEE152 pKa = 4.23NSTGSDD158 pKa = 3.43YY159 pKa = 11.23EE160 pKa = 4.07ANKK163 pKa = 10.68NIFRR167 pKa = 11.84SGPINSYY174 pKa = 9.95FQAEE178 pKa = 4.32AMGGYY183 pKa = 10.43ADD185 pKa = 4.54YY186 pKa = 11.26LSDD189 pKa = 3.27HH190 pKa = 6.77HH191 pKa = 6.6GWNTFAYY198 pKa = 10.14LADD201 pKa = 4.28DD202 pKa = 4.85AAWTDD207 pKa = 3.6PFTNNLPDD215 pKa = 3.61EE216 pKa = 4.69LEE218 pKa = 4.0SRR220 pKa = 11.84GYY222 pKa = 10.68DD223 pKa = 3.14VVYY226 pKa = 10.38EE227 pKa = 4.47SEE229 pKa = 4.57LSTGIDD235 pKa = 3.4DD236 pKa = 5.2FSTVMSDD243 pKa = 4.64LEE245 pKa = 4.42SEE247 pKa = 4.32EE248 pKa = 4.01PDD250 pKa = 3.13AVFRR254 pKa = 11.84FFAHH258 pKa = 7.54IIATDD263 pKa = 3.79MLASWHH269 pKa = 5.18QRR271 pKa = 11.84QAGFGIEE278 pKa = 4.55GIHH281 pKa = 5.67VASMTPDD288 pKa = 4.31FYY290 pKa = 11.42QLSEE294 pKa = 4.04GTATFEE300 pKa = 4.36TTSQSGAAGTTAITDD315 pKa = 3.51KK316 pKa = 10.95TLDD319 pKa = 3.79FVDD322 pKa = 5.08EE323 pKa = 4.32YY324 pKa = 11.59QAFTEE329 pKa = 4.96DD330 pKa = 5.03DD331 pKa = 4.32DD332 pKa = 4.71GAPDD336 pKa = 3.57LPMYY340 pKa = 10.22MGFNTYY346 pKa = 10.57DD347 pKa = 3.54AIYY350 pKa = 9.68LYY352 pKa = 10.79RR353 pKa = 11.84EE354 pKa = 4.17AVEE357 pKa = 4.22EE358 pKa = 4.54AGTADD363 pKa = 3.89YY364 pKa = 11.1EE365 pKa = 4.53SDD367 pKa = 4.13LDD369 pKa = 4.95DD370 pKa = 5.1IVDD373 pKa = 3.57AMLGLDD379 pKa = 3.6YY380 pKa = 10.83TGTAGEE386 pKa = 3.79ISFYY390 pKa = 11.37GEE392 pKa = 3.67GDD394 pKa = 3.83EE395 pKa = 4.88YY396 pKa = 10.8PHH398 pKa = 7.25DD399 pKa = 3.95VQEE402 pKa = 4.36EE403 pKa = 4.18RR404 pKa = 11.84GEE406 pKa = 4.11GDD408 pKa = 4.22AITNFPMTQWRR419 pKa = 11.84PEE421 pKa = 3.82GGLEE425 pKa = 4.07CVYY428 pKa = 10.6PEE430 pKa = 3.83QHH432 pKa = 5.63RR433 pKa = 11.84TADD436 pKa = 3.41HH437 pKa = 6.09VQPEE441 pKa = 4.38WMSS444 pKa = 3.25
MM1 pKa = 7.7ANDD4 pKa = 4.68DD5 pKa = 4.12NNTSKK10 pKa = 9.2TCRR13 pKa = 11.84IGKK16 pKa = 7.86TSVGRR21 pKa = 11.84RR22 pKa = 11.84TFLGAAGAGAVATTLAGCLGGGDD45 pKa = 4.52DD46 pKa = 5.23DD47 pKa = 5.43GFTIGHH53 pKa = 7.6LAPLEE58 pKa = 4.02NTQGLGSEE66 pKa = 4.47QSAEE70 pKa = 3.95LAAKK74 pKa = 9.33EE75 pKa = 4.15INDD78 pKa = 3.93DD79 pKa = 3.6GGIRR83 pKa = 11.84DD84 pKa = 3.93EE85 pKa = 4.41DD86 pKa = 4.12VEE88 pKa = 4.43IVSADD93 pKa = 3.48TRR95 pKa = 11.84SDD97 pKa = 3.2PSTAQDD103 pKa = 3.29EE104 pKa = 4.46ASRR107 pKa = 11.84LINQEE112 pKa = 3.82RR113 pKa = 11.84VDD115 pKa = 4.12LLVGTFSSEE124 pKa = 3.85VSVNIMDD131 pKa = 5.01MIAEE135 pKa = 4.07NDD137 pKa = 3.17VPYY140 pKa = 10.31IVTGSASPAIIEE152 pKa = 4.23NSTGSDD158 pKa = 3.43YY159 pKa = 11.23EE160 pKa = 4.07ANKK163 pKa = 10.68NIFRR167 pKa = 11.84SGPINSYY174 pKa = 9.95FQAEE178 pKa = 4.32AMGGYY183 pKa = 10.43ADD185 pKa = 4.54YY186 pKa = 11.26LSDD189 pKa = 3.27HH190 pKa = 6.77HH191 pKa = 6.6GWNTFAYY198 pKa = 10.14LADD201 pKa = 4.28DD202 pKa = 4.85AAWTDD207 pKa = 3.6PFTNNLPDD215 pKa = 3.61EE216 pKa = 4.69LEE218 pKa = 4.0SRR220 pKa = 11.84GYY222 pKa = 10.68DD223 pKa = 3.14VVYY226 pKa = 10.38EE227 pKa = 4.47SEE229 pKa = 4.57LSTGIDD235 pKa = 3.4DD236 pKa = 5.2FSTVMSDD243 pKa = 4.64LEE245 pKa = 4.42SEE247 pKa = 4.32EE248 pKa = 4.01PDD250 pKa = 3.13AVFRR254 pKa = 11.84FFAHH258 pKa = 7.54IIATDD263 pKa = 3.79MLASWHH269 pKa = 5.18QRR271 pKa = 11.84QAGFGIEE278 pKa = 4.55GIHH281 pKa = 5.67VASMTPDD288 pKa = 4.31FYY290 pKa = 11.42QLSEE294 pKa = 4.04GTATFEE300 pKa = 4.36TTSQSGAAGTTAITDD315 pKa = 3.51KK316 pKa = 10.95TLDD319 pKa = 3.79FVDD322 pKa = 5.08EE323 pKa = 4.32YY324 pKa = 11.59QAFTEE329 pKa = 4.96DD330 pKa = 5.03DD331 pKa = 4.32DD332 pKa = 4.71GAPDD336 pKa = 3.57LPMYY340 pKa = 10.22MGFNTYY346 pKa = 10.57DD347 pKa = 3.54AIYY350 pKa = 9.68LYY352 pKa = 10.79RR353 pKa = 11.84EE354 pKa = 4.17AVEE357 pKa = 4.22EE358 pKa = 4.54AGTADD363 pKa = 3.89YY364 pKa = 11.1EE365 pKa = 4.53SDD367 pKa = 4.13LDD369 pKa = 4.95DD370 pKa = 5.1IVDD373 pKa = 3.57AMLGLDD379 pKa = 3.6YY380 pKa = 10.83TGTAGEE386 pKa = 3.79ISFYY390 pKa = 11.37GEE392 pKa = 3.67GDD394 pKa = 3.83EE395 pKa = 4.88YY396 pKa = 10.8PHH398 pKa = 7.25DD399 pKa = 3.95VQEE402 pKa = 4.36EE403 pKa = 4.18RR404 pKa = 11.84GEE406 pKa = 4.11GDD408 pKa = 4.22AITNFPMTQWRR419 pKa = 11.84PEE421 pKa = 3.82GGLEE425 pKa = 4.07CVYY428 pKa = 10.6PEE430 pKa = 3.83QHH432 pKa = 5.63RR433 pKa = 11.84TADD436 pKa = 3.41HH437 pKa = 6.09VQPEE441 pKa = 4.38WMSS444 pKa = 3.25
Molecular weight: 48.21 kDa
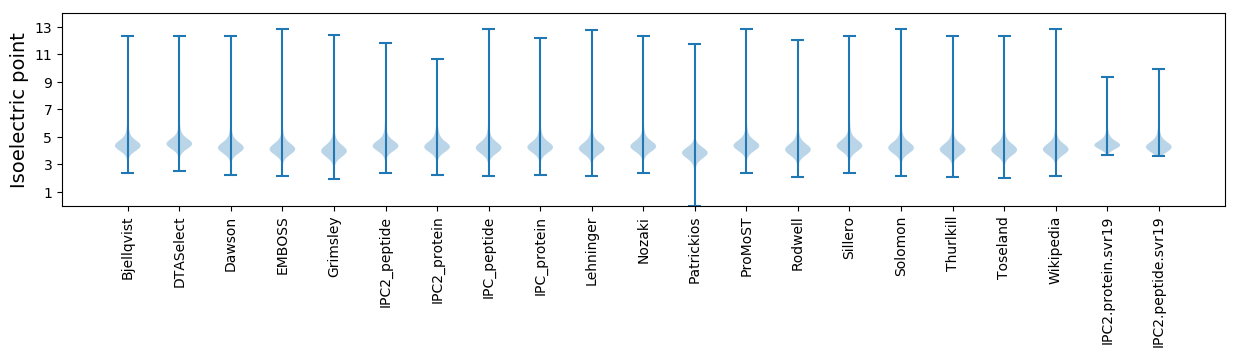
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I0MMJ3|A0A1I0MMJ3_9EURY Uncharacterized protein OS=Halopiger salifodinae OX=1202768 GN=SAMN05216285_1118 PE=4 SV=1
MM1 pKa = 8.29RR2 pKa = 11.84YY3 pKa = 9.14RR4 pKa = 11.84EE5 pKa = 3.64RR6 pKa = 11.84SIRR9 pKa = 11.84RR10 pKa = 11.84GQRR13 pKa = 11.84RR14 pKa = 11.84QHH16 pKa = 3.58VHH18 pKa = 5.57KK19 pKa = 10.57RR20 pKa = 11.84LAEE23 pKa = 3.98LEE25 pKa = 4.27RR26 pKa = 11.84EE27 pKa = 4.25LEE29 pKa = 4.11EE30 pKa = 3.69TRR32 pKa = 11.84DD33 pKa = 3.32RR34 pKa = 11.84VRR36 pKa = 11.84QLEE39 pKa = 4.03NTLRR43 pKa = 11.84GVVQNANDD51 pKa = 3.48VSIGGPCKK59 pKa = 10.2CGKK62 pKa = 9.91SLLLIRR68 pKa = 11.84QQKK71 pKa = 9.4IYY73 pKa = 10.48CPHH76 pKa = 6.73CGYY79 pKa = 10.58NRR81 pKa = 11.84TMM83 pKa = 4.06
MM1 pKa = 8.29RR2 pKa = 11.84YY3 pKa = 9.14RR4 pKa = 11.84EE5 pKa = 3.64RR6 pKa = 11.84SIRR9 pKa = 11.84RR10 pKa = 11.84GQRR13 pKa = 11.84RR14 pKa = 11.84QHH16 pKa = 3.58VHH18 pKa = 5.57KK19 pKa = 10.57RR20 pKa = 11.84LAEE23 pKa = 3.98LEE25 pKa = 4.27RR26 pKa = 11.84EE27 pKa = 4.25LEE29 pKa = 4.11EE30 pKa = 3.69TRR32 pKa = 11.84DD33 pKa = 3.32RR34 pKa = 11.84VRR36 pKa = 11.84QLEE39 pKa = 4.03NTLRR43 pKa = 11.84GVVQNANDD51 pKa = 3.48VSIGGPCKK59 pKa = 10.2CGKK62 pKa = 9.91SLLLIRR68 pKa = 11.84QQKK71 pKa = 9.4IYY73 pKa = 10.48CPHH76 pKa = 6.73CGYY79 pKa = 10.58NRR81 pKa = 11.84TMM83 pKa = 4.06
Molecular weight: 9.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1200987 |
29 |
1673 |
293.6 |
31.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.047 ± 0.051 | 0.754 ± 0.014 |
8.856 ± 0.047 | 8.89 ± 0.055 |
3.219 ± 0.025 | 8.478 ± 0.034 |
1.938 ± 0.019 | 4.333 ± 0.031 |
1.69 ± 0.02 | 8.744 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.712 ± 0.016 | 2.314 ± 0.021 |
4.741 ± 0.025 | 2.399 ± 0.027 |
6.647 ± 0.046 | 5.399 ± 0.029 |
6.336 ± 0.029 | 8.639 ± 0.038 |
1.143 ± 0.016 | 2.72 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
