
Aquila chrysaetos chrysaetos
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Deuterostomia; Chordata; Craniata; Vertebrata; Gnathostomata; Teleostomi; Euteleostomi; Sarcopterygii; Dipnotetrapodomorpha; Tetrapoda; Amniota; Sauropsida; Sauria; Archelosauria; Archosauria; Dinosauria; Saurischia; Theropoda;
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
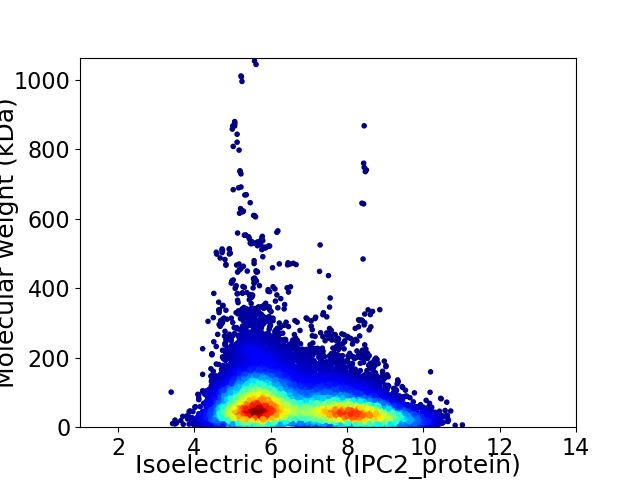
Virtual 2D-PAGE plot for 24817 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A663EC83|A0A663EC83_AQUCH Mannosyltransferase OS=Aquila chrysaetos chrysaetos OX=223781 GN=PIGB PE=3 SV=1
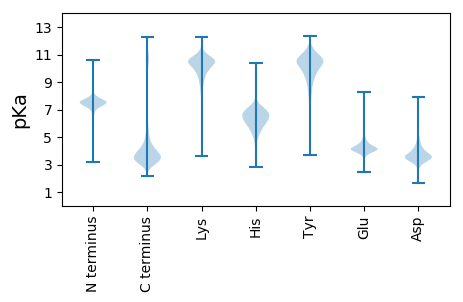
MM1 pKa = 7.97DD2 pKa = 4.61KK3 pKa = 10.51CTDD6 pKa = 3.5TEE8 pKa = 4.09EE9 pKa = 4.46DD10 pKa = 3.55QLLEE14 pKa = 4.41EE15 pKa = 5.27PDD17 pKa = 3.17IQYY20 pKa = 10.81SSKK23 pKa = 9.05VTQHH27 pKa = 6.28PSIASSIAEE36 pKa = 4.36SKK38 pKa = 10.7SSPGSDD44 pKa = 2.91GASSPEE50 pKa = 4.06GPEE53 pKa = 4.08LVYY56 pKa = 10.66IPAKK60 pKa = 9.4PAQLAAHH67 pKa = 5.94VLGNTDD73 pKa = 2.2SFYY76 pKa = 11.09SVRR79 pKa = 11.84DD80 pKa = 3.41VATSVQTLHH89 pKa = 7.14EE90 pKa = 4.8DD91 pKa = 3.46SQTSEE96 pKa = 4.2NEE98 pKa = 4.02CTPVEE103 pKa = 4.58GAAEE107 pKa = 4.36DD108 pKa = 4.47PSAVPAADD116 pKa = 3.41ASVEE120 pKa = 4.09AVTPQPGAWSQSSIAGEE137 pKa = 4.34LGPSDD142 pKa = 3.62SQADD146 pKa = 3.61VSTNDD151 pKa = 2.67VDD153 pKa = 3.87QASLVSLWEE162 pKa = 4.64DD163 pKa = 3.76PSPLPLSLPPSPTPKK178 pKa = 10.55DD179 pKa = 3.35PLQAVPSCNEE189 pKa = 3.77AEE191 pKa = 4.43VTSTTEE197 pKa = 4.14VVSLCWDD204 pKa = 3.89DD205 pKa = 4.42EE206 pKa = 5.03LIMDD210 pKa = 5.26AGVLPYY216 pKa = 10.35VEE218 pKa = 5.19QFPQKK223 pKa = 10.38IIIPFVDD230 pKa = 3.32QTAYY234 pKa = 10.73LLKK237 pKa = 10.13IEE239 pKa = 4.26QPLNVGQGLSATASGSSDD257 pKa = 3.73DD258 pKa = 5.84DD259 pKa = 4.06LVCHH263 pKa = 6.42PEE265 pKa = 3.99RR266 pKa = 11.84TEE268 pKa = 3.76STVQVEE274 pKa = 4.41SAEE277 pKa = 4.11QVYY280 pKa = 10.64VMSGKK285 pKa = 10.05KK286 pKa = 7.88VWVFVSSSCFSFQLYY301 pKa = 10.74SFISGHH307 pKa = 5.53
MM1 pKa = 7.97DD2 pKa = 4.61KK3 pKa = 10.51CTDD6 pKa = 3.5TEE8 pKa = 4.09EE9 pKa = 4.46DD10 pKa = 3.55QLLEE14 pKa = 4.41EE15 pKa = 5.27PDD17 pKa = 3.17IQYY20 pKa = 10.81SSKK23 pKa = 9.05VTQHH27 pKa = 6.28PSIASSIAEE36 pKa = 4.36SKK38 pKa = 10.7SSPGSDD44 pKa = 2.91GASSPEE50 pKa = 4.06GPEE53 pKa = 4.08LVYY56 pKa = 10.66IPAKK60 pKa = 9.4PAQLAAHH67 pKa = 5.94VLGNTDD73 pKa = 2.2SFYY76 pKa = 11.09SVRR79 pKa = 11.84DD80 pKa = 3.41VATSVQTLHH89 pKa = 7.14EE90 pKa = 4.8DD91 pKa = 3.46SQTSEE96 pKa = 4.2NEE98 pKa = 4.02CTPVEE103 pKa = 4.58GAAEE107 pKa = 4.36DD108 pKa = 4.47PSAVPAADD116 pKa = 3.41ASVEE120 pKa = 4.09AVTPQPGAWSQSSIAGEE137 pKa = 4.34LGPSDD142 pKa = 3.62SQADD146 pKa = 3.61VSTNDD151 pKa = 2.67VDD153 pKa = 3.87QASLVSLWEE162 pKa = 4.64DD163 pKa = 3.76PSPLPLSLPPSPTPKK178 pKa = 10.55DD179 pKa = 3.35PLQAVPSCNEE189 pKa = 3.77AEE191 pKa = 4.43VTSTTEE197 pKa = 4.14VVSLCWDD204 pKa = 3.89DD205 pKa = 4.42EE206 pKa = 5.03LIMDD210 pKa = 5.26AGVLPYY216 pKa = 10.35VEE218 pKa = 5.19QFPQKK223 pKa = 10.38IIIPFVDD230 pKa = 3.32QTAYY234 pKa = 10.73LLKK237 pKa = 10.13IEE239 pKa = 4.26QPLNVGQGLSATASGSSDD257 pKa = 3.73DD258 pKa = 5.84DD259 pKa = 4.06LVCHH263 pKa = 6.42PEE265 pKa = 3.99RR266 pKa = 11.84TEE268 pKa = 3.76STVQVEE274 pKa = 4.41SAEE277 pKa = 4.11QVYY280 pKa = 10.64VMSGKK285 pKa = 10.05KK286 pKa = 7.88VWVFVSSSCFSFQLYY301 pKa = 10.74SFISGHH307 pKa = 5.53
Molecular weight: 32.6 kDa
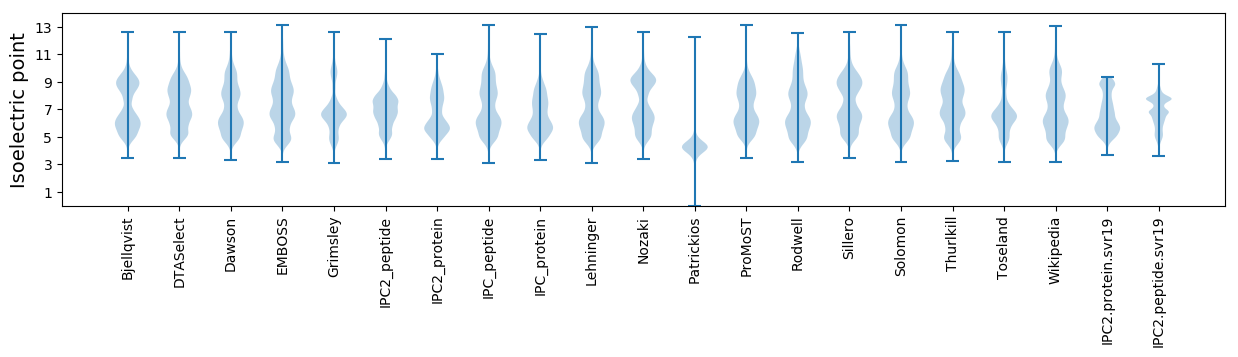
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A663ER34|A0A663ER34_AQUCH Transmembrane protein 222 OS=Aquila chrysaetos chrysaetos OX=223781 GN=TMEM222 PE=4 SV=1
MM1 pKa = 8.1DD2 pKa = 5.03PRR4 pKa = 11.84APSALTVPDD13 pKa = 4.43PPSPEE18 pKa = 4.19SPRR21 pKa = 11.84PAHH24 pKa = 6.43GPGPHH29 pKa = 6.33VSPPGPSAPSPATRR43 pKa = 11.84QQGRR47 pKa = 11.84SPAPHH52 pKa = 6.12SPARR56 pKa = 11.84PWAPRR61 pKa = 11.84GQAHH65 pKa = 7.13PWAHH69 pKa = 6.84LPPVGPSPARR79 pKa = 11.84GPIISRR85 pKa = 11.84PWAHH89 pKa = 7.05LPPVGPSPAHH99 pKa = 5.96GPIISRR105 pKa = 11.84PWAHH109 pKa = 7.05LPPVGPSPTHH119 pKa = 6.21GPISRR124 pKa = 11.84PWAHH128 pKa = 6.85HH129 pKa = 6.43LPPVGPSPARR139 pKa = 11.84GPIISRR145 pKa = 11.84PWAHH149 pKa = 6.82HH150 pKa = 6.43LPPVGPSPARR160 pKa = 11.84GPISHH165 pKa = 7.58PWAHH169 pKa = 6.95LPPVGPSSPARR180 pKa = 11.84GPISHH185 pKa = 7.58PWAHH189 pKa = 6.95LPPVGPSSPARR200 pKa = 11.84GPISRR205 pKa = 11.84PWAHH209 pKa = 6.85HH210 pKa = 6.43LPPVGPSPTRR220 pKa = 11.84GPISRR225 pKa = 11.84PWAHH229 pKa = 6.85HH230 pKa = 6.43LPPVGPSSPARR241 pKa = 11.84GPISRR246 pKa = 11.84PWAHH250 pKa = 6.85HH251 pKa = 6.43LPPVGPSPARR261 pKa = 11.84GPISHH266 pKa = 7.58PWAHH270 pKa = 6.95LPPVGPSSPARR281 pKa = 11.84GPISHH286 pKa = 7.58PWAHH290 pKa = 6.95LPPVGPSSPARR301 pKa = 11.84GPIISRR307 pKa = 11.84PWAHH311 pKa = 7.05LPPVGPSPGLAPSPRR326 pKa = 11.84RR327 pKa = 11.84CPPPVPP333 pKa = 3.91
MM1 pKa = 8.1DD2 pKa = 5.03PRR4 pKa = 11.84APSALTVPDD13 pKa = 4.43PPSPEE18 pKa = 4.19SPRR21 pKa = 11.84PAHH24 pKa = 6.43GPGPHH29 pKa = 6.33VSPPGPSAPSPATRR43 pKa = 11.84QQGRR47 pKa = 11.84SPAPHH52 pKa = 6.12SPARR56 pKa = 11.84PWAPRR61 pKa = 11.84GQAHH65 pKa = 7.13PWAHH69 pKa = 6.84LPPVGPSPARR79 pKa = 11.84GPIISRR85 pKa = 11.84PWAHH89 pKa = 7.05LPPVGPSPAHH99 pKa = 5.96GPIISRR105 pKa = 11.84PWAHH109 pKa = 7.05LPPVGPSPTHH119 pKa = 6.21GPISRR124 pKa = 11.84PWAHH128 pKa = 6.85HH129 pKa = 6.43LPPVGPSPARR139 pKa = 11.84GPIISRR145 pKa = 11.84PWAHH149 pKa = 6.82HH150 pKa = 6.43LPPVGPSPARR160 pKa = 11.84GPISHH165 pKa = 7.58PWAHH169 pKa = 6.95LPPVGPSSPARR180 pKa = 11.84GPISHH185 pKa = 7.58PWAHH189 pKa = 6.95LPPVGPSSPARR200 pKa = 11.84GPISRR205 pKa = 11.84PWAHH209 pKa = 6.85HH210 pKa = 6.43LPPVGPSPTRR220 pKa = 11.84GPISRR225 pKa = 11.84PWAHH229 pKa = 6.85HH230 pKa = 6.43LPPVGPSSPARR241 pKa = 11.84GPISRR246 pKa = 11.84PWAHH250 pKa = 6.85HH251 pKa = 6.43LPPVGPSPARR261 pKa = 11.84GPISHH266 pKa = 7.58PWAHH270 pKa = 6.95LPPVGPSSPARR281 pKa = 11.84GPISHH286 pKa = 7.58PWAHH290 pKa = 6.95LPPVGPSSPARR301 pKa = 11.84GPIISRR307 pKa = 11.84PWAHH311 pKa = 7.05LPPVGPSPGLAPSPRR326 pKa = 11.84RR327 pKa = 11.84CPPPVPP333 pKa = 3.91
Molecular weight: 34.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
16571618 |
20 |
9572 |
667.8 |
74.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.905 ± 0.014 | 2.234 ± 0.014 |
4.985 ± 0.009 | 7.268 ± 0.02 |
3.623 ± 0.01 | 6.226 ± 0.02 |
2.514 ± 0.007 | 4.648 ± 0.011 |
6.101 ± 0.019 | 9.672 ± 0.018 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.208 ± 0.006 | 3.924 ± 0.01 |
5.715 ± 0.019 | 4.755 ± 0.013 |
5.494 ± 0.013 | 8.276 ± 0.017 |
5.338 ± 0.01 | 6.127 ± 0.011 |
1.191 ± 0.005 | 2.794 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
