
Streptococcus satellite phage Javan352
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.9
Get precalculated fractions of proteins
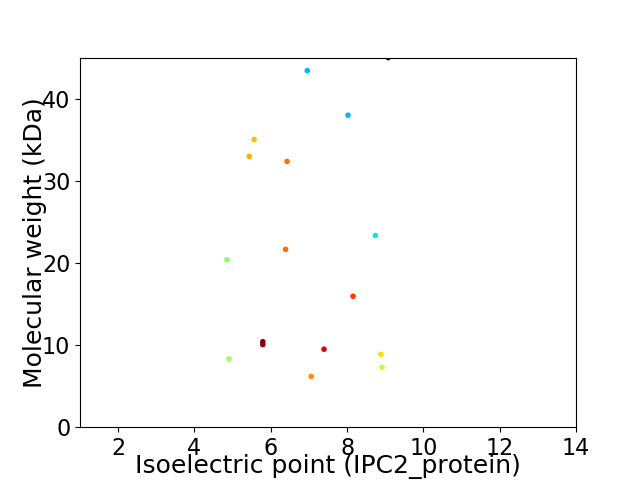
Virtual 2D-PAGE plot for 17 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4D5ZKA2|A0A4D5ZKA2_9VIRU Uncharacterized protein OS=Streptococcus satellite phage Javan352 OX=2558653 GN=JavanS352_0008 PE=4 SV=1
MM1 pKa = 7.6EE2 pKa = 6.11KK3 pKa = 10.91NMTLDD8 pKa = 3.77NMTQSEE14 pKa = 4.5FDD16 pKa = 3.49NRR18 pKa = 11.84ITEE21 pKa = 4.12IKK23 pKa = 10.3DD24 pKa = 3.2RR25 pKa = 11.84NPNLFQFIIDD35 pKa = 3.88FLDD38 pKa = 4.06DD39 pKa = 3.54KK40 pKa = 10.4VTPEE44 pKa = 3.89EE45 pKa = 4.34VYY47 pKa = 11.05DD48 pKa = 3.84FLKK51 pKa = 10.18MEE53 pKa = 4.85RR54 pKa = 11.84SYY56 pKa = 11.18QVNYY60 pKa = 9.19IKK62 pKa = 10.55NYY64 pKa = 8.5KK65 pKa = 9.6ARR67 pKa = 11.84AA68 pKa = 3.49
MM1 pKa = 7.6EE2 pKa = 6.11KK3 pKa = 10.91NMTLDD8 pKa = 3.77NMTQSEE14 pKa = 4.5FDD16 pKa = 3.49NRR18 pKa = 11.84ITEE21 pKa = 4.12IKK23 pKa = 10.3DD24 pKa = 3.2RR25 pKa = 11.84NPNLFQFIIDD35 pKa = 3.88FLDD38 pKa = 4.06DD39 pKa = 3.54KK40 pKa = 10.4VTPEE44 pKa = 3.89EE45 pKa = 4.34VYY47 pKa = 11.05DD48 pKa = 3.84FLKK51 pKa = 10.18MEE53 pKa = 4.85RR54 pKa = 11.84SYY56 pKa = 11.18QVNYY60 pKa = 9.19IKK62 pKa = 10.55NYY64 pKa = 8.5KK65 pKa = 9.6ARR67 pKa = 11.84AA68 pKa = 3.49
Molecular weight: 8.32 kDa
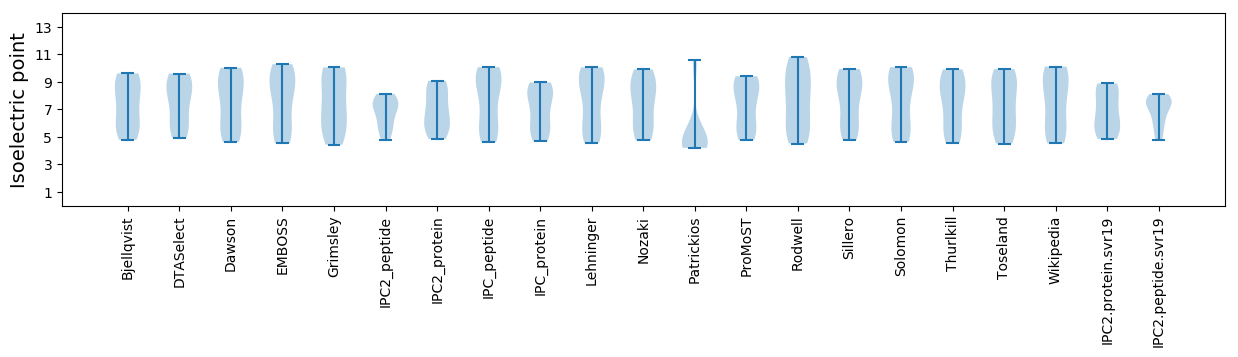
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4D5ZQM5|A0A4D5ZQM5_9VIRU Uncharacterized protein OS=Streptococcus satellite phage Javan352 OX=2558653 GN=JavanS352_0003 PE=4 SV=1
MM1 pKa = 7.99KK2 pKa = 9.31ITQHH6 pKa = 4.84TKK8 pKa = 9.3KK9 pKa = 10.42DD10 pKa = 3.23GSAVYY15 pKa = 10.05RR16 pKa = 11.84SSIYY20 pKa = 10.71LGIDD24 pKa = 2.85SVTGKK29 pKa = 10.14KK30 pKa = 10.39VKK32 pKa = 8.21TTISARR38 pKa = 11.84TKK40 pKa = 10.43KK41 pKa = 9.83EE42 pKa = 3.68LRR44 pKa = 11.84NKK46 pKa = 8.17ATQAKK51 pKa = 10.07VEE53 pKa = 4.17FEE55 pKa = 4.59KK56 pKa = 11.24NGSTRR61 pKa = 11.84KK62 pKa = 9.17QRR64 pKa = 11.84SHH66 pKa = 5.33ITTYY70 pKa = 11.06SEE72 pKa = 4.48LVDD75 pKa = 5.66LFWQTYY81 pKa = 5.79QHH83 pKa = 6.36TIKK86 pKa = 10.07TNTQIKK92 pKa = 10.32IKK94 pKa = 10.47GCLKK98 pKa = 10.72NYY100 pKa = 9.18LLPSFSTYY108 pKa = 11.08KK109 pKa = 10.24LDD111 pKa = 3.42KK112 pKa = 8.8LTPVIIQTQVNKK124 pKa = 9.76WADD127 pKa = 3.8EE128 pKa = 4.09YY129 pKa = 11.47NQDD132 pKa = 2.92GTGYY136 pKa = 10.61KK137 pKa = 9.95EE138 pKa = 4.0YY139 pKa = 11.21NHH141 pKa = 6.5LHH143 pKa = 6.37ALNKK147 pKa = 10.25RR148 pKa = 11.84ILQYY152 pKa = 10.81GVSIQALDD160 pKa = 3.67NNPARR165 pKa = 11.84DD166 pKa = 3.34IVIPRR171 pKa = 11.84KK172 pKa = 8.2ITRR175 pKa = 11.84DD176 pKa = 3.35KK177 pKa = 11.58QEE179 pKa = 3.32IKK181 pKa = 10.77YY182 pKa = 9.97FQDD185 pKa = 3.26QEE187 pKa = 4.42LKK189 pKa = 11.12NFLSYY194 pKa = 11.28LDD196 pKa = 3.81NLEE199 pKa = 3.97NTFVNFYY206 pKa = 10.04DD207 pKa = 3.65TVLYY211 pKa = 11.0KK212 pKa = 10.08MLLATGLRR220 pKa = 11.84IRR222 pKa = 11.84EE223 pKa = 4.14CLALEE228 pKa = 4.16WSDD231 pKa = 4.72IDD233 pKa = 4.27LQNGTIDD240 pKa = 3.28INKK243 pKa = 8.44TLNILNQVNTPKK255 pKa = 10.11TKK257 pKa = 9.97SSYY260 pKa = 9.92RR261 pKa = 11.84VLDD264 pKa = 3.58IDD266 pKa = 3.85HH267 pKa = 6.69KK268 pKa = 8.91TVLMLRR274 pKa = 11.84LYY276 pKa = 10.46RR277 pKa = 11.84ARR279 pKa = 11.84QAEE282 pKa = 4.01NGRR285 pKa = 11.84NIGLTYY291 pKa = 10.56EE292 pKa = 4.48KK293 pKa = 10.64VFSDD297 pKa = 4.51SFDD300 pKa = 3.64NYY302 pKa = 10.22VNTRR306 pKa = 11.84KK307 pKa = 9.81VDD309 pKa = 3.68YY310 pKa = 10.13RR311 pKa = 11.84LHH313 pKa = 6.06KK314 pKa = 9.82HH315 pKa = 6.03LKK317 pKa = 9.47SANCTDD323 pKa = 3.87LGFHH327 pKa = 7.4AFRR330 pKa = 11.84HH331 pKa = 4.54THH333 pKa = 6.71ASILLNAGLPYY344 pKa = 10.43KK345 pKa = 10.2EE346 pKa = 3.68IQTRR350 pKa = 11.84LGHH353 pKa = 6.33AKK355 pKa = 10.05ISVTMDD361 pKa = 3.68TYY363 pKa = 11.66SHH365 pKa = 7.23LSKK368 pKa = 10.56EE369 pKa = 4.22NQKK372 pKa = 10.33RR373 pKa = 11.84AVSFFEE379 pKa = 4.04TALEE383 pKa = 4.69KK384 pKa = 10.71INSSS388 pKa = 3.32
MM1 pKa = 7.99KK2 pKa = 9.31ITQHH6 pKa = 4.84TKK8 pKa = 9.3KK9 pKa = 10.42DD10 pKa = 3.23GSAVYY15 pKa = 10.05RR16 pKa = 11.84SSIYY20 pKa = 10.71LGIDD24 pKa = 2.85SVTGKK29 pKa = 10.14KK30 pKa = 10.39VKK32 pKa = 8.21TTISARR38 pKa = 11.84TKK40 pKa = 10.43KK41 pKa = 9.83EE42 pKa = 3.68LRR44 pKa = 11.84NKK46 pKa = 8.17ATQAKK51 pKa = 10.07VEE53 pKa = 4.17FEE55 pKa = 4.59KK56 pKa = 11.24NGSTRR61 pKa = 11.84KK62 pKa = 9.17QRR64 pKa = 11.84SHH66 pKa = 5.33ITTYY70 pKa = 11.06SEE72 pKa = 4.48LVDD75 pKa = 5.66LFWQTYY81 pKa = 5.79QHH83 pKa = 6.36TIKK86 pKa = 10.07TNTQIKK92 pKa = 10.32IKK94 pKa = 10.47GCLKK98 pKa = 10.72NYY100 pKa = 9.18LLPSFSTYY108 pKa = 11.08KK109 pKa = 10.24LDD111 pKa = 3.42KK112 pKa = 8.8LTPVIIQTQVNKK124 pKa = 9.76WADD127 pKa = 3.8EE128 pKa = 4.09YY129 pKa = 11.47NQDD132 pKa = 2.92GTGYY136 pKa = 10.61KK137 pKa = 9.95EE138 pKa = 4.0YY139 pKa = 11.21NHH141 pKa = 6.5LHH143 pKa = 6.37ALNKK147 pKa = 10.25RR148 pKa = 11.84ILQYY152 pKa = 10.81GVSIQALDD160 pKa = 3.67NNPARR165 pKa = 11.84DD166 pKa = 3.34IVIPRR171 pKa = 11.84KK172 pKa = 8.2ITRR175 pKa = 11.84DD176 pKa = 3.35KK177 pKa = 11.58QEE179 pKa = 3.32IKK181 pKa = 10.77YY182 pKa = 9.97FQDD185 pKa = 3.26QEE187 pKa = 4.42LKK189 pKa = 11.12NFLSYY194 pKa = 11.28LDD196 pKa = 3.81NLEE199 pKa = 3.97NTFVNFYY206 pKa = 10.04DD207 pKa = 3.65TVLYY211 pKa = 11.0KK212 pKa = 10.08MLLATGLRR220 pKa = 11.84IRR222 pKa = 11.84EE223 pKa = 4.14CLALEE228 pKa = 4.16WSDD231 pKa = 4.72IDD233 pKa = 4.27LQNGTIDD240 pKa = 3.28INKK243 pKa = 8.44TLNILNQVNTPKK255 pKa = 10.11TKK257 pKa = 9.97SSYY260 pKa = 9.92RR261 pKa = 11.84VLDD264 pKa = 3.58IDD266 pKa = 3.85HH267 pKa = 6.69KK268 pKa = 8.91TVLMLRR274 pKa = 11.84LYY276 pKa = 10.46RR277 pKa = 11.84ARR279 pKa = 11.84QAEE282 pKa = 4.01NGRR285 pKa = 11.84NIGLTYY291 pKa = 10.56EE292 pKa = 4.48KK293 pKa = 10.64VFSDD297 pKa = 4.51SFDD300 pKa = 3.64NYY302 pKa = 10.22VNTRR306 pKa = 11.84KK307 pKa = 9.81VDD309 pKa = 3.68YY310 pKa = 10.13RR311 pKa = 11.84LHH313 pKa = 6.06KK314 pKa = 9.82HH315 pKa = 6.03LKK317 pKa = 9.47SANCTDD323 pKa = 3.87LGFHH327 pKa = 7.4AFRR330 pKa = 11.84HH331 pKa = 4.54THH333 pKa = 6.71ASILLNAGLPYY344 pKa = 10.43KK345 pKa = 10.2EE346 pKa = 3.68IQTRR350 pKa = 11.84LGHH353 pKa = 6.33AKK355 pKa = 10.05ISVTMDD361 pKa = 3.68TYY363 pKa = 11.66SHH365 pKa = 7.23LSKK368 pKa = 10.56EE369 pKa = 4.22NQKK372 pKa = 10.33RR373 pKa = 11.84AVSFFEE379 pKa = 4.04TALEE383 pKa = 4.69KK384 pKa = 10.71INSSS388 pKa = 3.32
Molecular weight: 45.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3160 |
53 |
388 |
185.9 |
21.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.861 ± 0.406 | 0.601 ± 0.096 |
6.266 ± 0.418 | 8.386 ± 0.645 |
5.19 ± 0.628 | 4.367 ± 0.537 |
1.677 ± 0.325 | 7.12 ± 0.424 |
10.854 ± 0.522 | 10.316 ± 0.372 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.057 ± 0.278 | 5.981 ± 0.417 |
2.057 ± 0.244 | 3.987 ± 0.349 |
4.652 ± 0.336 | 6.329 ± 0.436 |
6.234 ± 0.577 | 4.842 ± 0.424 |
0.633 ± 0.119 | 4.589 ± 0.315 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
