
Torque teno zalophus virus 1 (ZcTTV)
Taxonomy: Viruses; Anelloviridae; Lambdatorquevirus; Torque teno pinniped virus 5
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
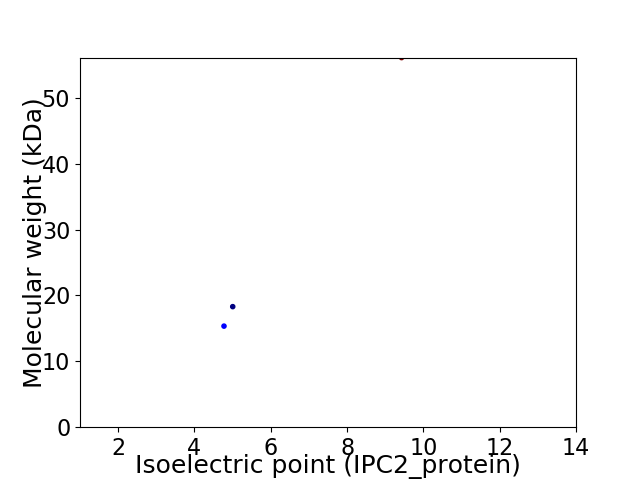
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
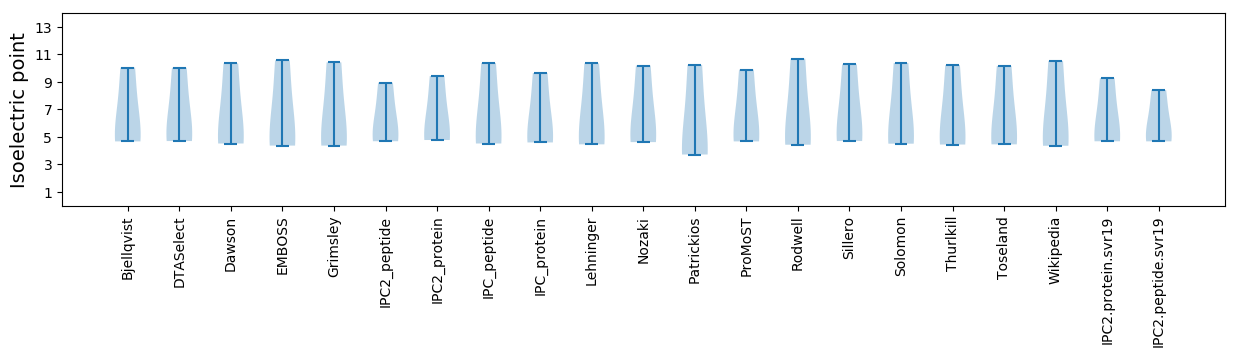
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C0JSK8|C0JSK8_ZCTTV Capsid protein OS=Torque teno zalophus virus 1 OX=1218490 PE=3 SV=1
MM1 pKa = 7.72EE2 pKa = 6.87DD3 pKa = 4.76IRR5 pKa = 11.84TLWKK9 pKa = 9.31VTFDD13 pKa = 4.21LLYY16 pKa = 10.73DD17 pKa = 3.63NNYY20 pKa = 9.52FPSTGMYY27 pKa = 9.48PCFDD31 pKa = 3.59EE32 pKa = 6.91KK33 pKa = 11.03EE34 pKa = 4.09ICLCLAWTLLLIGFSTLLILTIQLIISVRR63 pKa = 11.84RR64 pKa = 11.84LNGSVNALISIKK76 pKa = 10.6NSAGVVTGQTISPKK90 pKa = 9.83NAISEE95 pKa = 4.24EE96 pKa = 4.25VQAPEE101 pKa = 4.08MEE103 pKa = 4.34EE104 pKa = 3.96TLQEE108 pKa = 4.23VPLSEE113 pKa = 4.24EE114 pKa = 4.21SPLVGAQTQKK124 pKa = 10.07MASPTKK130 pKa = 9.54NACGKK135 pKa = 10.08IPSFF139 pKa = 4.32
MM1 pKa = 7.72EE2 pKa = 6.87DD3 pKa = 4.76IRR5 pKa = 11.84TLWKK9 pKa = 9.31VTFDD13 pKa = 4.21LLYY16 pKa = 10.73DD17 pKa = 3.63NNYY20 pKa = 9.52FPSTGMYY27 pKa = 9.48PCFDD31 pKa = 3.59EE32 pKa = 6.91KK33 pKa = 11.03EE34 pKa = 4.09ICLCLAWTLLLIGFSTLLILTIQLIISVRR63 pKa = 11.84RR64 pKa = 11.84LNGSVNALISIKK76 pKa = 10.6NSAGVVTGQTISPKK90 pKa = 9.83NAISEE95 pKa = 4.24EE96 pKa = 4.25VQAPEE101 pKa = 4.08MEE103 pKa = 4.34EE104 pKa = 3.96TLQEE108 pKa = 4.23VPLSEE113 pKa = 4.24EE114 pKa = 4.21SPLVGAQTQKK124 pKa = 10.07MASPTKK130 pKa = 9.54NACGKK135 pKa = 10.08IPSFF139 pKa = 4.32
Molecular weight: 15.34 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C0JSK9|C0JSK9_ZCTTV Orf3 OS=Torque teno zalophus virus 1 OX=1218490 PE=4 SV=1
MM1 pKa = 7.66PFRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84FRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84WRR13 pKa = 11.84RR14 pKa = 11.84PFRR17 pKa = 11.84RR18 pKa = 11.84YY19 pKa = 8.63HH20 pKa = 4.7YY21 pKa = 10.11RR22 pKa = 11.84RR23 pKa = 11.84NRR25 pKa = 11.84HH26 pKa = 2.89WWGRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84KK34 pKa = 7.62WRR36 pKa = 11.84HH37 pKa = 4.46RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84MPAVRR45 pKa = 11.84YY46 pKa = 9.21HH47 pKa = 6.37PSRR50 pKa = 11.84RR51 pKa = 11.84HH52 pKa = 4.91KK53 pKa = 10.46YY54 pKa = 8.39IVIRR58 pKa = 11.84GIEE61 pKa = 4.05PLGNLCTDD69 pKa = 4.36HH70 pKa = 6.35TPSWLRR76 pKa = 11.84ATPGRR81 pKa = 11.84SFEE84 pKa = 4.56SAGQTLGEE92 pKa = 4.27WEE94 pKa = 4.56GTWGAHH100 pKa = 4.21HH101 pKa = 6.92HH102 pKa = 6.23SFAGLLLRR110 pKa = 11.84AQCRR114 pKa = 11.84FATFSGDD121 pKa = 2.86WKK123 pKa = 10.58SYY125 pKa = 11.12DD126 pKa = 3.46YY127 pKa = 11.03IEE129 pKa = 4.42YY130 pKa = 10.48KK131 pKa = 10.77GGTFYY136 pKa = 10.66IPPQCTTFLFGIDD149 pKa = 3.54PQFTKK154 pKa = 10.35ISKK157 pKa = 9.86EE158 pKa = 4.14GEE160 pKa = 3.82KK161 pKa = 10.2EE162 pKa = 4.06QPNEE166 pKa = 4.2EE167 pKa = 4.25TWLHH171 pKa = 6.03PGWLLHH177 pKa = 6.16QRR179 pKa = 11.84GTHH182 pKa = 6.25IIYY185 pKa = 10.87SKK187 pKa = 10.44DD188 pKa = 2.9IKK190 pKa = 10.13PWRR193 pKa = 11.84RR194 pKa = 11.84WYY196 pKa = 10.45KK197 pKa = 10.65LRR199 pKa = 11.84VKK201 pKa = 10.31PGPTWEE207 pKa = 4.83GPYY210 pKa = 10.35SLPNAFNFIMSQWWWSWLDD229 pKa = 3.49FQHH232 pKa = 7.16AFEE235 pKa = 6.04DD236 pKa = 3.75NTRR239 pKa = 11.84SQICHH244 pKa = 7.2ADD246 pKa = 3.47PQNFSLFCGQKK257 pKa = 9.56PWWFDD262 pKa = 3.07STYY265 pKa = 10.72KK266 pKa = 9.46NTIEE270 pKa = 3.91IPKK273 pKa = 9.93RR274 pKa = 11.84LVNKK278 pKa = 10.11ACNAWVNRR286 pKa = 11.84QEE288 pKa = 3.89YY289 pKa = 8.69MIKK292 pKa = 9.84IGQKK296 pKa = 8.49VTDD299 pKa = 4.03KK300 pKa = 10.31MLHH303 pKa = 6.27PEE305 pKa = 4.66DD306 pKa = 6.37DD307 pKa = 3.94ILCGNQSQQSQKK319 pKa = 11.09KK320 pKa = 9.14YY321 pKa = 10.18FQQQNPGWGPFLPLLYY337 pKa = 11.01NGDD340 pKa = 3.91NCSLWFKK347 pKa = 10.25YY348 pKa = 10.13RR349 pKa = 11.84YY350 pKa = 8.45VFKK353 pKa = 11.23VSGDD357 pKa = 3.36CEE359 pKa = 4.18YY360 pKa = 11.14RR361 pKa = 11.84KK362 pKa = 8.04TPSTDD367 pKa = 2.87LTQMVPTAPGPWNAEE382 pKa = 3.34GDD384 pKa = 3.78RR385 pKa = 11.84PQIQSRR391 pKa = 11.84SILKK395 pKa = 10.22KK396 pKa = 9.43SRR398 pKa = 11.84KK399 pKa = 9.29RR400 pKa = 11.84PLDD403 pKa = 3.34TADD406 pKa = 3.71ILLGDD411 pKa = 4.11TDD413 pKa = 5.57DD414 pKa = 6.14GILTEE419 pKa = 4.52TGLEE423 pKa = 4.24RR424 pKa = 11.84ISGPSPKK431 pKa = 10.28DD432 pKa = 3.3LLCGMEE438 pKa = 4.04NTPPKK443 pKa = 9.96RR444 pKa = 11.84VRR446 pKa = 11.84FRR448 pKa = 11.84QSDD451 pKa = 3.74VLRR454 pKa = 11.84KK455 pKa = 9.37HH456 pKa = 5.66KK457 pKa = 10.3HH458 pKa = 5.95RR459 pKa = 11.84ILNLIDD465 pKa = 3.27QLSRR469 pKa = 3.35
MM1 pKa = 7.66PFRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84FRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84RR11 pKa = 11.84WRR13 pKa = 11.84RR14 pKa = 11.84PFRR17 pKa = 11.84RR18 pKa = 11.84YY19 pKa = 8.63HH20 pKa = 4.7YY21 pKa = 10.11RR22 pKa = 11.84RR23 pKa = 11.84NRR25 pKa = 11.84HH26 pKa = 2.89WWGRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84KK34 pKa = 7.62WRR36 pKa = 11.84HH37 pKa = 4.46RR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84MPAVRR45 pKa = 11.84YY46 pKa = 9.21HH47 pKa = 6.37PSRR50 pKa = 11.84RR51 pKa = 11.84HH52 pKa = 4.91KK53 pKa = 10.46YY54 pKa = 8.39IVIRR58 pKa = 11.84GIEE61 pKa = 4.05PLGNLCTDD69 pKa = 4.36HH70 pKa = 6.35TPSWLRR76 pKa = 11.84ATPGRR81 pKa = 11.84SFEE84 pKa = 4.56SAGQTLGEE92 pKa = 4.27WEE94 pKa = 4.56GTWGAHH100 pKa = 4.21HH101 pKa = 6.92HH102 pKa = 6.23SFAGLLLRR110 pKa = 11.84AQCRR114 pKa = 11.84FATFSGDD121 pKa = 2.86WKK123 pKa = 10.58SYY125 pKa = 11.12DD126 pKa = 3.46YY127 pKa = 11.03IEE129 pKa = 4.42YY130 pKa = 10.48KK131 pKa = 10.77GGTFYY136 pKa = 10.66IPPQCTTFLFGIDD149 pKa = 3.54PQFTKK154 pKa = 10.35ISKK157 pKa = 9.86EE158 pKa = 4.14GEE160 pKa = 3.82KK161 pKa = 10.2EE162 pKa = 4.06QPNEE166 pKa = 4.2EE167 pKa = 4.25TWLHH171 pKa = 6.03PGWLLHH177 pKa = 6.16QRR179 pKa = 11.84GTHH182 pKa = 6.25IIYY185 pKa = 10.87SKK187 pKa = 10.44DD188 pKa = 2.9IKK190 pKa = 10.13PWRR193 pKa = 11.84RR194 pKa = 11.84WYY196 pKa = 10.45KK197 pKa = 10.65LRR199 pKa = 11.84VKK201 pKa = 10.31PGPTWEE207 pKa = 4.83GPYY210 pKa = 10.35SLPNAFNFIMSQWWWSWLDD229 pKa = 3.49FQHH232 pKa = 7.16AFEE235 pKa = 6.04DD236 pKa = 3.75NTRR239 pKa = 11.84SQICHH244 pKa = 7.2ADD246 pKa = 3.47PQNFSLFCGQKK257 pKa = 9.56PWWFDD262 pKa = 3.07STYY265 pKa = 10.72KK266 pKa = 9.46NTIEE270 pKa = 3.91IPKK273 pKa = 9.93RR274 pKa = 11.84LVNKK278 pKa = 10.11ACNAWVNRR286 pKa = 11.84QEE288 pKa = 3.89YY289 pKa = 8.69MIKK292 pKa = 9.84IGQKK296 pKa = 8.49VTDD299 pKa = 4.03KK300 pKa = 10.31MLHH303 pKa = 6.27PEE305 pKa = 4.66DD306 pKa = 6.37DD307 pKa = 3.94ILCGNQSQQSQKK319 pKa = 11.09KK320 pKa = 9.14YY321 pKa = 10.18FQQQNPGWGPFLPLLYY337 pKa = 11.01NGDD340 pKa = 3.91NCSLWFKK347 pKa = 10.25YY348 pKa = 10.13RR349 pKa = 11.84YY350 pKa = 8.45VFKK353 pKa = 11.23VSGDD357 pKa = 3.36CEE359 pKa = 4.18YY360 pKa = 11.14RR361 pKa = 11.84KK362 pKa = 8.04TPSTDD367 pKa = 2.87LTQMVPTAPGPWNAEE382 pKa = 3.34GDD384 pKa = 3.78RR385 pKa = 11.84PQIQSRR391 pKa = 11.84SILKK395 pKa = 10.22KK396 pKa = 9.43SRR398 pKa = 11.84KK399 pKa = 9.29RR400 pKa = 11.84PLDD403 pKa = 3.34TADD406 pKa = 3.71ILLGDD411 pKa = 4.11TDD413 pKa = 5.57DD414 pKa = 6.14GILTEE419 pKa = 4.52TGLEE423 pKa = 4.24RR424 pKa = 11.84ISGPSPKK431 pKa = 10.28DD432 pKa = 3.3LLCGMEE438 pKa = 4.04NTPPKK443 pKa = 9.96RR444 pKa = 11.84VRR446 pKa = 11.84FRR448 pKa = 11.84QSDD451 pKa = 3.74VLRR454 pKa = 11.84KK455 pKa = 9.37HH456 pKa = 5.66KK457 pKa = 10.3HH458 pKa = 5.95RR459 pKa = 11.84ILNLIDD465 pKa = 3.27QLSRR469 pKa = 3.35
Molecular weight: 56.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
770 |
139 |
469 |
256.7 |
29.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.636 ± 0.582 | 1.948 ± 0.435 |
4.545 ± 0.532 | 5.974 ± 1.467 |
4.026 ± 0.814 | 5.844 ± 0.867 |
3.117 ± 0.854 | 5.325 ± 0.985 |
5.714 ± 0.593 | 8.182 ± 1.107 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.948 ± 0.386 | 3.896 ± 0.345 |
6.494 ± 0.658 | 5.974 ± 1.049 |
7.532 ± 2.624 | 8.182 ± 2.359 |
7.143 ± 1.483 | 3.506 ± 0.978 |
3.506 ± 1.176 | 3.506 ± 0.379 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
