
bacterium K02(2017)
Taxonomy: cellular organisms; Bacteria; unclassified Bacteria
Average proteome isoelectric point is 6.91
Get precalculated fractions of proteins
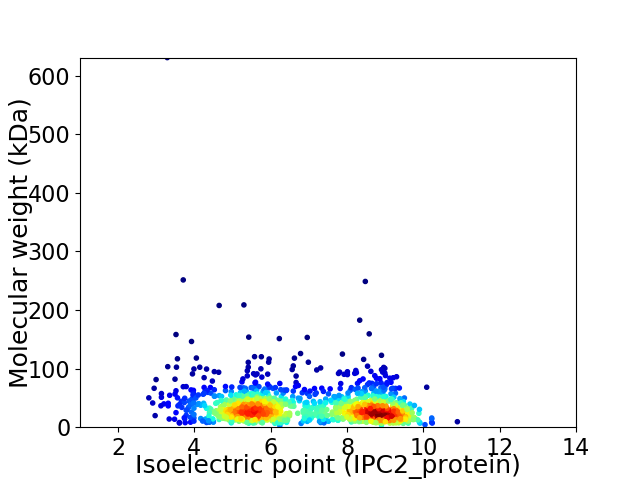
Virtual 2D-PAGE plot for 1213 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A202E270|A0A202E270_9BACT S4 RNA-binding domain-containing protein OS=bacterium K02(2017) OX=1932696 GN=BVY03_01135 PE=4 SV=1
MM1 pKa = 7.11VPGVKK6 pKa = 9.88KK7 pKa = 10.64ISFLFITLLVLLVGSLVVLAADD29 pKa = 5.02IIDD32 pKa = 4.0DD33 pKa = 4.12TEE35 pKa = 5.19AEE37 pKa = 4.33FDD39 pKa = 3.35NGSYY43 pKa = 10.86TNVDD47 pKa = 3.98FDD49 pKa = 4.42TGNTWVEE56 pKa = 3.95LTAPATNAYY65 pKa = 10.42GNFEE69 pKa = 4.14SDD71 pKa = 3.54IYY73 pKa = 11.43DD74 pKa = 3.61SGGTATWNTISWVPNRR90 pKa = 11.84PSGKK94 pKa = 9.48QLLNDD99 pKa = 4.13GGTEE103 pKa = 4.04GSSYY107 pKa = 10.92DD108 pKa = 3.42INNISMSGNVSVWHH122 pKa = 6.31FNASSWNGTAGEE134 pKa = 4.27VVDD137 pKa = 6.27DD138 pKa = 4.55NDD140 pKa = 3.79ASEE143 pKa = 4.13NAHH146 pKa = 6.86DD147 pKa = 4.45GVSVTSVTTDD157 pKa = 2.87STGMFNTRR165 pKa = 11.84SAEE168 pKa = 3.76FDD170 pKa = 3.54DD171 pKa = 5.19DD172 pKa = 4.2GGRR175 pKa = 11.84LEE177 pKa = 4.97IADD180 pKa = 4.66HH181 pKa = 6.77NDD183 pKa = 2.71FDD185 pKa = 4.91ASSFTLEE192 pKa = 3.28AWVYY196 pKa = 10.16PKK198 pKa = 9.53STPDD202 pKa = 3.14WNVVAIKK209 pKa = 9.22ATNDD213 pKa = 3.2AFDD216 pKa = 5.44DD217 pKa = 4.0GWGMVIQGSEE227 pKa = 3.86VKK229 pKa = 10.47FYY231 pKa = 11.43VQDD234 pKa = 3.49QANYY238 pKa = 8.18SASIAVTTNEE248 pKa = 3.73WNHH251 pKa = 5.81IVGTYY256 pKa = 10.18DD257 pKa = 3.13GSNVRR262 pKa = 11.84SYY264 pKa = 11.61INGTIDD270 pKa = 3.13STVAYY275 pKa = 7.24TGPYY279 pKa = 10.03DD280 pKa = 3.81GTSTALMIGGDD291 pKa = 3.69NFYY294 pKa = 11.22DD295 pKa = 3.65SDD297 pKa = 3.62TWDD300 pKa = 3.66GYY302 pKa = 9.64IDD304 pKa = 3.68EE305 pKa = 4.46VAFYY309 pKa = 11.04DD310 pKa = 3.88VVLDD314 pKa = 4.08AGEE317 pKa = 3.69INARR321 pKa = 11.84YY322 pKa = 9.39KK323 pKa = 10.71RR324 pKa = 11.84GIQRR328 pKa = 11.84VRR330 pKa = 11.84FQVRR334 pKa = 11.84SCDD337 pKa = 3.56DD338 pKa = 3.7AACSGEE344 pKa = 4.24NFEE347 pKa = 5.67GPDD350 pKa = 3.65GTTGTYY356 pKa = 10.64YY357 pKa = 10.64SASSNLITATPTATLTNVDD376 pKa = 3.81DD377 pKa = 4.24NQYY380 pKa = 9.47FQYY383 pKa = 10.85KK384 pKa = 9.93IFFEE388 pKa = 4.34TDD390 pKa = 3.0DD391 pKa = 3.69TGLTPEE397 pKa = 4.41INSVTVNYY405 pKa = 10.61DD406 pKa = 3.33LVSSDD411 pKa = 4.06FVSSKK416 pKa = 10.59QKK418 pKa = 10.47ISDD421 pKa = 3.51TAGVFTAVLDD431 pKa = 3.67NSDD434 pKa = 3.79YY435 pKa = 11.39FGVTISNIGDD445 pKa = 3.97LDD447 pKa = 3.89GDD449 pKa = 4.65SINDD453 pKa = 4.66LIVGAHH459 pKa = 5.87QDD461 pKa = 3.64DD462 pKa = 4.82DD463 pKa = 4.35GGADD467 pKa = 3.28RR468 pKa = 11.84GAVYY472 pKa = 10.89VLFLEE477 pKa = 4.43SDD479 pKa = 3.85GTVSSFQKK487 pKa = 10.59ISDD490 pKa = 3.7TAGSFTAVLDD500 pKa = 3.55NTDD503 pKa = 3.29YY504 pKa = 11.33FGISAASIGDD514 pKa = 3.49IDD516 pKa = 3.99TDD518 pKa = 4.58GINDD522 pKa = 3.49IAVGAFFDD530 pKa = 5.99DD531 pKa = 5.16DD532 pKa = 4.01GGTNKK537 pKa = 10.22GAVYY541 pKa = 10.42ILFLEE546 pKa = 4.27NDD548 pKa = 3.92GTVSSFQKK556 pKa = 10.59ISDD559 pKa = 3.7TAGSFTASFTVTDD572 pKa = 4.04LFGRR576 pKa = 11.84SVASMGDD583 pKa = 3.5VNGDD587 pKa = 3.95SVPDD591 pKa = 3.67IAVGAYY597 pKa = 9.94YY598 pKa = 10.58DD599 pKa = 4.84DD600 pKa = 6.12DD601 pKa = 5.74GGTDD605 pKa = 2.98RR606 pKa = 11.84GAVYY610 pKa = 10.25IIFLNNDD617 pKa = 3.19GTVKK621 pKa = 10.43SFQKK625 pKa = 10.62ISDD628 pKa = 3.94TEE630 pKa = 4.41GSFTGVLDD638 pKa = 3.42NTDD641 pKa = 3.38YY642 pKa = 11.35FGVSVASIKK651 pKa = 10.56DD652 pKa = 3.68LNGDD656 pKa = 4.67GINEE660 pKa = 4.28LAVGSYY666 pKa = 10.94YY667 pKa = 10.85DD668 pKa = 4.56DD669 pKa = 5.45DD670 pKa = 5.3GGSEE674 pKa = 3.9RR675 pKa = 11.84GAVYY679 pKa = 11.23VMFLNNNGTVSSFQKK694 pKa = 10.6ISDD697 pKa = 3.91TEE699 pKa = 4.36GSFTAVLDD707 pKa = 3.68NTNGFGVSVDD717 pKa = 3.64GLNDD721 pKa = 3.48FDD723 pKa = 6.23GDD725 pKa = 4.42GITDD729 pKa = 3.95LMVGAHH735 pKa = 6.54SDD737 pKa = 3.59NDD739 pKa = 3.71GGTDD743 pKa = 3.04NGAIYY748 pKa = 10.68VLYY751 pKa = 10.6LNANGTVSSYY761 pKa = 11.37EE762 pKa = 4.19KK763 pKa = 10.57ISDD766 pKa = 3.78TEE768 pKa = 4.3GGFTGVLDD776 pKa = 3.59NSDD779 pKa = 3.67YY780 pKa = 11.35FGISAANIADD790 pKa = 4.1LDD792 pKa = 4.22GDD794 pKa = 4.38GVTDD798 pKa = 3.81IAIGAYY804 pKa = 9.97GDD806 pKa = 3.8ADD808 pKa = 3.52GGLNRR813 pKa = 11.84GAVHH817 pKa = 7.23ILNISQPDD825 pKa = 3.94SYY827 pKa = 11.48SVGSITINAQMAINRR842 pKa = 11.84NTNNANTKK850 pKa = 7.73VTITFTIAGAIPVNGDD866 pKa = 2.4IDD868 pKa = 4.42IIFPTGFILTEE879 pKa = 4.02VGSGDD884 pKa = 3.53VSEE887 pKa = 5.05ADD889 pKa = 3.34SDD891 pKa = 4.12GGTLTTSVSGQRR903 pKa = 11.84MTINCGSSITASTEE917 pKa = 3.6LALTIDD923 pKa = 3.76NVVNSASAGDD933 pKa = 3.94YY934 pKa = 10.09EE935 pKa = 4.41GLIIEE940 pKa = 4.63TQDD943 pKa = 3.21ASDD946 pKa = 3.75VVIDD950 pKa = 4.15LVEE953 pKa = 5.01SDD955 pKa = 4.07GVTVIQKK962 pKa = 8.98PRR964 pKa = 11.84RR965 pKa = 11.84ILTSQQ970 pKa = 2.86
MM1 pKa = 7.11VPGVKK6 pKa = 9.88KK7 pKa = 10.64ISFLFITLLVLLVGSLVVLAADD29 pKa = 5.02IIDD32 pKa = 4.0DD33 pKa = 4.12TEE35 pKa = 5.19AEE37 pKa = 4.33FDD39 pKa = 3.35NGSYY43 pKa = 10.86TNVDD47 pKa = 3.98FDD49 pKa = 4.42TGNTWVEE56 pKa = 3.95LTAPATNAYY65 pKa = 10.42GNFEE69 pKa = 4.14SDD71 pKa = 3.54IYY73 pKa = 11.43DD74 pKa = 3.61SGGTATWNTISWVPNRR90 pKa = 11.84PSGKK94 pKa = 9.48QLLNDD99 pKa = 4.13GGTEE103 pKa = 4.04GSSYY107 pKa = 10.92DD108 pKa = 3.42INNISMSGNVSVWHH122 pKa = 6.31FNASSWNGTAGEE134 pKa = 4.27VVDD137 pKa = 6.27DD138 pKa = 4.55NDD140 pKa = 3.79ASEE143 pKa = 4.13NAHH146 pKa = 6.86DD147 pKa = 4.45GVSVTSVTTDD157 pKa = 2.87STGMFNTRR165 pKa = 11.84SAEE168 pKa = 3.76FDD170 pKa = 3.54DD171 pKa = 5.19DD172 pKa = 4.2GGRR175 pKa = 11.84LEE177 pKa = 4.97IADD180 pKa = 4.66HH181 pKa = 6.77NDD183 pKa = 2.71FDD185 pKa = 4.91ASSFTLEE192 pKa = 3.28AWVYY196 pKa = 10.16PKK198 pKa = 9.53STPDD202 pKa = 3.14WNVVAIKK209 pKa = 9.22ATNDD213 pKa = 3.2AFDD216 pKa = 5.44DD217 pKa = 4.0GWGMVIQGSEE227 pKa = 3.86VKK229 pKa = 10.47FYY231 pKa = 11.43VQDD234 pKa = 3.49QANYY238 pKa = 8.18SASIAVTTNEE248 pKa = 3.73WNHH251 pKa = 5.81IVGTYY256 pKa = 10.18DD257 pKa = 3.13GSNVRR262 pKa = 11.84SYY264 pKa = 11.61INGTIDD270 pKa = 3.13STVAYY275 pKa = 7.24TGPYY279 pKa = 10.03DD280 pKa = 3.81GTSTALMIGGDD291 pKa = 3.69NFYY294 pKa = 11.22DD295 pKa = 3.65SDD297 pKa = 3.62TWDD300 pKa = 3.66GYY302 pKa = 9.64IDD304 pKa = 3.68EE305 pKa = 4.46VAFYY309 pKa = 11.04DD310 pKa = 3.88VVLDD314 pKa = 4.08AGEE317 pKa = 3.69INARR321 pKa = 11.84YY322 pKa = 9.39KK323 pKa = 10.71RR324 pKa = 11.84GIQRR328 pKa = 11.84VRR330 pKa = 11.84FQVRR334 pKa = 11.84SCDD337 pKa = 3.56DD338 pKa = 3.7AACSGEE344 pKa = 4.24NFEE347 pKa = 5.67GPDD350 pKa = 3.65GTTGTYY356 pKa = 10.64YY357 pKa = 10.64SASSNLITATPTATLTNVDD376 pKa = 3.81DD377 pKa = 4.24NQYY380 pKa = 9.47FQYY383 pKa = 10.85KK384 pKa = 9.93IFFEE388 pKa = 4.34TDD390 pKa = 3.0DD391 pKa = 3.69TGLTPEE397 pKa = 4.41INSVTVNYY405 pKa = 10.61DD406 pKa = 3.33LVSSDD411 pKa = 4.06FVSSKK416 pKa = 10.59QKK418 pKa = 10.47ISDD421 pKa = 3.51TAGVFTAVLDD431 pKa = 3.67NSDD434 pKa = 3.79YY435 pKa = 11.39FGVTISNIGDD445 pKa = 3.97LDD447 pKa = 3.89GDD449 pKa = 4.65SINDD453 pKa = 4.66LIVGAHH459 pKa = 5.87QDD461 pKa = 3.64DD462 pKa = 4.82DD463 pKa = 4.35GGADD467 pKa = 3.28RR468 pKa = 11.84GAVYY472 pKa = 10.89VLFLEE477 pKa = 4.43SDD479 pKa = 3.85GTVSSFQKK487 pKa = 10.59ISDD490 pKa = 3.7TAGSFTAVLDD500 pKa = 3.55NTDD503 pKa = 3.29YY504 pKa = 11.33FGISAASIGDD514 pKa = 3.49IDD516 pKa = 3.99TDD518 pKa = 4.58GINDD522 pKa = 3.49IAVGAFFDD530 pKa = 5.99DD531 pKa = 5.16DD532 pKa = 4.01GGTNKK537 pKa = 10.22GAVYY541 pKa = 10.42ILFLEE546 pKa = 4.27NDD548 pKa = 3.92GTVSSFQKK556 pKa = 10.59ISDD559 pKa = 3.7TAGSFTASFTVTDD572 pKa = 4.04LFGRR576 pKa = 11.84SVASMGDD583 pKa = 3.5VNGDD587 pKa = 3.95SVPDD591 pKa = 3.67IAVGAYY597 pKa = 9.94YY598 pKa = 10.58DD599 pKa = 4.84DD600 pKa = 6.12DD601 pKa = 5.74GGTDD605 pKa = 2.98RR606 pKa = 11.84GAVYY610 pKa = 10.25IIFLNNDD617 pKa = 3.19GTVKK621 pKa = 10.43SFQKK625 pKa = 10.62ISDD628 pKa = 3.94TEE630 pKa = 4.41GSFTGVLDD638 pKa = 3.42NTDD641 pKa = 3.38YY642 pKa = 11.35FGVSVASIKK651 pKa = 10.56DD652 pKa = 3.68LNGDD656 pKa = 4.67GINEE660 pKa = 4.28LAVGSYY666 pKa = 10.94YY667 pKa = 10.85DD668 pKa = 4.56DD669 pKa = 5.45DD670 pKa = 5.3GGSEE674 pKa = 3.9RR675 pKa = 11.84GAVYY679 pKa = 11.23VMFLNNNGTVSSFQKK694 pKa = 10.6ISDD697 pKa = 3.91TEE699 pKa = 4.36GSFTAVLDD707 pKa = 3.68NTNGFGVSVDD717 pKa = 3.64GLNDD721 pKa = 3.48FDD723 pKa = 6.23GDD725 pKa = 4.42GITDD729 pKa = 3.95LMVGAHH735 pKa = 6.54SDD737 pKa = 3.59NDD739 pKa = 3.71GGTDD743 pKa = 3.04NGAIYY748 pKa = 10.68VLYY751 pKa = 10.6LNANGTVSSYY761 pKa = 11.37EE762 pKa = 4.19KK763 pKa = 10.57ISDD766 pKa = 3.78TEE768 pKa = 4.3GGFTGVLDD776 pKa = 3.59NSDD779 pKa = 3.67YY780 pKa = 11.35FGISAANIADD790 pKa = 4.1LDD792 pKa = 4.22GDD794 pKa = 4.38GVTDD798 pKa = 3.81IAIGAYY804 pKa = 9.97GDD806 pKa = 3.8ADD808 pKa = 3.52GGLNRR813 pKa = 11.84GAVHH817 pKa = 7.23ILNISQPDD825 pKa = 3.94SYY827 pKa = 11.48SVGSITINAQMAINRR842 pKa = 11.84NTNNANTKK850 pKa = 7.73VTITFTIAGAIPVNGDD866 pKa = 2.4IDD868 pKa = 4.42IIFPTGFILTEE879 pKa = 4.02VGSGDD884 pKa = 3.53VSEE887 pKa = 5.05ADD889 pKa = 3.34SDD891 pKa = 4.12GGTLTTSVSGQRR903 pKa = 11.84MTINCGSSITASTEE917 pKa = 3.6LALTIDD923 pKa = 3.76NVVNSASAGDD933 pKa = 3.94YY934 pKa = 10.09EE935 pKa = 4.41GLIIEE940 pKa = 4.63TQDD943 pKa = 3.21ASDD946 pKa = 3.75VVIDD950 pKa = 4.15LVEE953 pKa = 5.01SDD955 pKa = 4.07GVTVIQKK962 pKa = 8.98PRR964 pKa = 11.84RR965 pKa = 11.84ILTSQQ970 pKa = 2.86
Molecular weight: 102.42 kDa
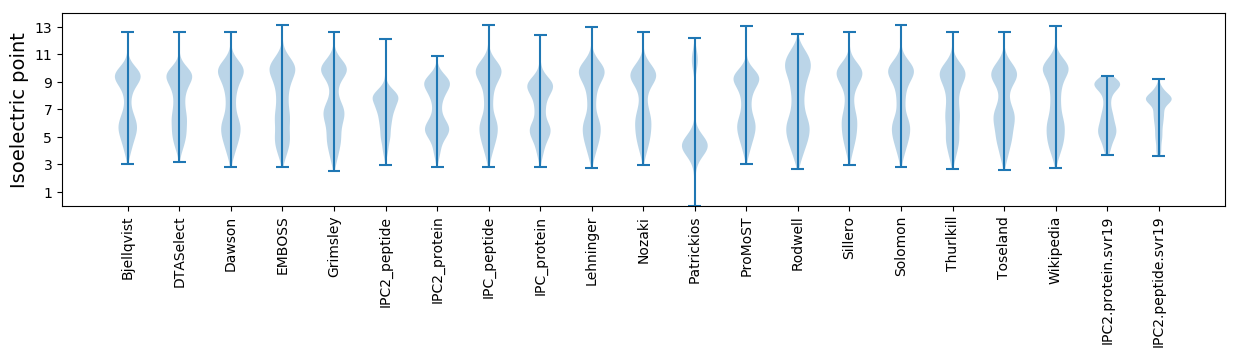
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A202DZ42|A0A202DZ42_9BACT Replicative DNA helicase OS=bacterium K02(2017) OX=1932696 GN=BVY03_03785 PE=3 SV=1
MM1 pKa = 7.79KK2 pKa = 10.07FRR4 pKa = 11.84KK5 pKa = 9.62KK6 pKa = 10.52LLSKK10 pKa = 10.69QKK12 pKa = 10.24LRR14 pKa = 11.84PKK16 pKa = 10.48LRR18 pKa = 11.84VKK20 pKa = 10.14QRR22 pKa = 11.84QKK24 pKa = 10.55QRR26 pKa = 11.84PKK28 pKa = 9.84LRR30 pKa = 11.84VRR32 pKa = 11.84QRR34 pKa = 11.84VRR36 pKa = 11.84QRR38 pKa = 11.84QKK40 pKa = 8.62PKK42 pKa = 10.03LRR44 pKa = 11.84VRR46 pKa = 11.84QRR48 pKa = 11.84QKK50 pKa = 10.61QKK52 pKa = 10.62QKK54 pKa = 10.97LRR56 pKa = 11.84LKK58 pKa = 10.69LKK60 pKa = 10.25LRR62 pKa = 11.84HH63 pKa = 5.51LQNLFVEE70 pKa = 4.89MEE72 pKa = 3.99NN73 pKa = 3.82
MM1 pKa = 7.79KK2 pKa = 10.07FRR4 pKa = 11.84KK5 pKa = 9.62KK6 pKa = 10.52LLSKK10 pKa = 10.69QKK12 pKa = 10.24LRR14 pKa = 11.84PKK16 pKa = 10.48LRR18 pKa = 11.84VKK20 pKa = 10.14QRR22 pKa = 11.84QKK24 pKa = 10.55QRR26 pKa = 11.84PKK28 pKa = 9.84LRR30 pKa = 11.84VRR32 pKa = 11.84QRR34 pKa = 11.84VRR36 pKa = 11.84QRR38 pKa = 11.84QKK40 pKa = 8.62PKK42 pKa = 10.03LRR44 pKa = 11.84VRR46 pKa = 11.84QRR48 pKa = 11.84QKK50 pKa = 10.61QKK52 pKa = 10.62QKK54 pKa = 10.97LRR56 pKa = 11.84LKK58 pKa = 10.69LKK60 pKa = 10.25LRR62 pKa = 11.84HH63 pKa = 5.51LQNLFVEE70 pKa = 4.89MEE72 pKa = 3.99NN73 pKa = 3.82
Molecular weight: 9.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
401908 |
24 |
6113 |
331.3 |
37.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.494 ± 0.082 | 0.997 ± 0.035 |
5.875 ± 0.084 | 5.778 ± 0.068 |
4.712 ± 0.07 | 6.293 ± 0.085 |
2.037 ± 0.042 | 8.148 ± 0.133 |
7.719 ± 0.145 | 9.888 ± 0.093 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.103 ± 0.04 | 6.022 ± 0.079 |
3.57 ± 0.061 | 3.988 ± 0.058 |
3.443 ± 0.065 | 7.026 ± 0.075 |
5.548 ± 0.122 | 5.886 ± 0.058 |
0.927 ± 0.025 | 3.544 ± 0.059 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
