
Ustilaginoidea virens RNA virus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; Victorivirus; unclassified Victorivirus
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
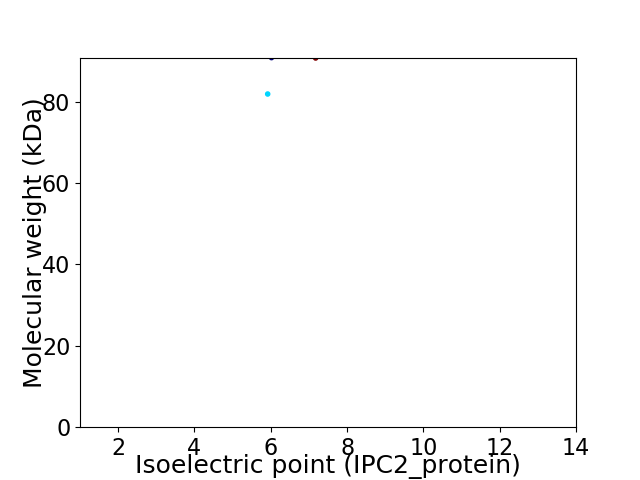
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M9QS71|M9QS71_9VIRU Putative coat protein OS=Ustilaginoidea virens RNA virus 1 OX=1312445 PE=4 SV=1
MM1 pKa = 7.34SALSTFLSGGVVAKK15 pKa = 10.88GPLKK19 pKa = 9.87NNEE22 pKa = 3.24VRR24 pKa = 11.84RR25 pKa = 11.84YY26 pKa = 6.98YY27 pKa = 8.24TTLISEE33 pKa = 4.17ATLNGVQDD41 pKa = 3.83VAGKK45 pKa = 7.52TVVWKK50 pKa = 10.67VGYY53 pKa = 9.9AYY55 pKa = 10.73SGTAGIDD62 pKa = 3.18YY63 pKa = 10.79DD64 pKa = 4.19GVYY67 pKa = 10.04KK68 pKa = 10.56IHH70 pKa = 6.69AEE72 pKa = 4.08KK73 pKa = 10.9VRR75 pKa = 11.84ADD77 pKa = 3.46PKK79 pKa = 10.8AYY81 pKa = 9.64PNSKK85 pKa = 9.67KK86 pKa = 10.37RR87 pKa = 11.84NEE89 pKa = 4.06LLKK92 pKa = 10.43PLIRR96 pKa = 11.84EE97 pKa = 4.43AYY99 pKa = 8.33NLKK102 pKa = 10.66DD103 pKa = 3.84SEE105 pKa = 4.39EE106 pKa = 4.31LPIPAALVCPKK117 pKa = 10.27SVAIRR122 pKa = 11.84LQTPYY127 pKa = 9.93PVNEE131 pKa = 4.72AISAEE136 pKa = 4.07FANAARR142 pKa = 11.84RR143 pKa = 11.84YY144 pKa = 8.36SGVVGTHH151 pKa = 5.46GTADD155 pKa = 3.21FCGIVAQLAKK165 pKa = 10.52GLAFFASTGGLTMRR179 pKa = 11.84DD180 pKa = 3.27LAGGNSYY187 pKa = 11.29SYY189 pKa = 9.98MAVGNHH195 pKa = 5.33TAPLVASTTSIWVPRR210 pKa = 11.84YY211 pKa = 9.92AEE213 pKa = 4.33SLMAPNIMAALVAAGSAVGSTIVTDD238 pKa = 4.5LLPTDD243 pKa = 3.75VTNTPLLTEE252 pKa = 4.47ANHH255 pKa = 6.62LEE257 pKa = 4.21LAAGVYY263 pKa = 9.56HH264 pKa = 7.51ALRR267 pKa = 11.84VLGTNMEE274 pKa = 4.42ANGAGQLFAYY284 pKa = 9.7AVTVGIHH291 pKa = 5.33SVVTVVGMTDD301 pKa = 2.73EE302 pKa = 4.44GAYY305 pKa = 9.32MRR307 pKa = 11.84DD308 pKa = 3.47VFRR311 pKa = 11.84STSFAPSYY319 pKa = 11.02GGISTVLPEE328 pKa = 3.73WHH330 pKa = 6.7GFPRR334 pKa = 11.84PADD337 pKa = 3.38SSLAGWVGLVDD348 pKa = 5.48SIALGTAASVAIADD362 pKa = 3.92PCVAIADD369 pKa = 4.31RR370 pKa = 11.84YY371 pKa = 10.67YY372 pKa = 9.48PTTLTGRR379 pKa = 11.84YY380 pKa = 8.77EE381 pKa = 4.16YY382 pKa = 10.94GGEE385 pKa = 3.88PGMGYY390 pKa = 10.38VGDD393 pKa = 4.26EE394 pKa = 3.66NDD396 pKa = 3.41ARR398 pKa = 11.84IISGKK403 pKa = 9.96IMQLAPSFTRR413 pKa = 11.84NYY415 pKa = 9.92FKK417 pKa = 11.13VLARR421 pKa = 11.84LFSVTPHH428 pKa = 6.34EE429 pKa = 4.39NSSVGLTHH437 pKa = 7.13MIHH440 pKa = 6.61AFSSEE445 pKa = 3.89TLRR448 pKa = 11.84DD449 pKa = 3.69SRR451 pKa = 11.84HH452 pKa = 5.41LQHH455 pKa = 6.99PSVSPFYY462 pKa = 9.42WVEE465 pKa = 3.5PTGIITFDD473 pKa = 3.36TSDD476 pKa = 3.62FTATAAGFGTLATPSQPGTIPMFEE500 pKa = 4.07RR501 pKa = 11.84AEE503 pKa = 4.14VTQNVGDD510 pKa = 3.88VSDD513 pKa = 4.64VLVAWRR519 pKa = 11.84SARR522 pKa = 11.84TSGLAIHH529 pKa = 6.92LNNHH533 pKa = 5.15VEE535 pKa = 4.09AGMEE539 pKa = 4.29NIRR542 pKa = 11.84ITGCDD547 pKa = 3.59PNSWAQLGGASQSVSNRR564 pKa = 11.84VTAGNDD570 pKa = 2.93LASYY574 pKa = 8.2MWARR578 pKa = 11.84SDD580 pKa = 3.56VGVPAPGEE588 pKa = 3.73AMYY591 pKa = 10.69LGEE594 pKa = 5.39AISLLVRR601 pKa = 11.84HH602 pKa = 6.32DD603 pKa = 3.96TLDD606 pKa = 3.54GNTWANTPNHH616 pKa = 5.92VPRR619 pKa = 11.84DD620 pKa = 3.71VEE622 pKa = 4.07FGNNVTIRR630 pKa = 11.84VGLLAPYY637 pKa = 9.96GVGPIGPNRR646 pKa = 11.84KK647 pKa = 9.44ANRR650 pKa = 11.84AFTAALRR657 pKa = 11.84ALDD660 pKa = 3.75AARR663 pKa = 11.84EE664 pKa = 4.22SGMIRR669 pKa = 11.84APKK672 pKa = 10.1LCGTIKK678 pKa = 10.29LTDD681 pKa = 3.37QAFGRR686 pKa = 11.84QAAVPVAIAPPSPKK700 pKa = 10.04AANRR704 pKa = 11.84PVTTVSQPPQSDD716 pKa = 3.25ARR718 pKa = 11.84PVEE721 pKa = 3.98PQNYY725 pKa = 9.03AEE727 pKa = 4.9SPTAHH732 pKa = 7.06VYY734 pKa = 8.2TQGPKK739 pKa = 8.66TAPGPAQSVIASVRR753 pKa = 11.84QMAAEE758 pKa = 4.31EE759 pKa = 4.2SAEE762 pKa = 4.27TASVAAAPQAGAAQQ776 pKa = 3.55
MM1 pKa = 7.34SALSTFLSGGVVAKK15 pKa = 10.88GPLKK19 pKa = 9.87NNEE22 pKa = 3.24VRR24 pKa = 11.84RR25 pKa = 11.84YY26 pKa = 6.98YY27 pKa = 8.24TTLISEE33 pKa = 4.17ATLNGVQDD41 pKa = 3.83VAGKK45 pKa = 7.52TVVWKK50 pKa = 10.67VGYY53 pKa = 9.9AYY55 pKa = 10.73SGTAGIDD62 pKa = 3.18YY63 pKa = 10.79DD64 pKa = 4.19GVYY67 pKa = 10.04KK68 pKa = 10.56IHH70 pKa = 6.69AEE72 pKa = 4.08KK73 pKa = 10.9VRR75 pKa = 11.84ADD77 pKa = 3.46PKK79 pKa = 10.8AYY81 pKa = 9.64PNSKK85 pKa = 9.67KK86 pKa = 10.37RR87 pKa = 11.84NEE89 pKa = 4.06LLKK92 pKa = 10.43PLIRR96 pKa = 11.84EE97 pKa = 4.43AYY99 pKa = 8.33NLKK102 pKa = 10.66DD103 pKa = 3.84SEE105 pKa = 4.39EE106 pKa = 4.31LPIPAALVCPKK117 pKa = 10.27SVAIRR122 pKa = 11.84LQTPYY127 pKa = 9.93PVNEE131 pKa = 4.72AISAEE136 pKa = 4.07FANAARR142 pKa = 11.84RR143 pKa = 11.84YY144 pKa = 8.36SGVVGTHH151 pKa = 5.46GTADD155 pKa = 3.21FCGIVAQLAKK165 pKa = 10.52GLAFFASTGGLTMRR179 pKa = 11.84DD180 pKa = 3.27LAGGNSYY187 pKa = 11.29SYY189 pKa = 9.98MAVGNHH195 pKa = 5.33TAPLVASTTSIWVPRR210 pKa = 11.84YY211 pKa = 9.92AEE213 pKa = 4.33SLMAPNIMAALVAAGSAVGSTIVTDD238 pKa = 4.5LLPTDD243 pKa = 3.75VTNTPLLTEE252 pKa = 4.47ANHH255 pKa = 6.62LEE257 pKa = 4.21LAAGVYY263 pKa = 9.56HH264 pKa = 7.51ALRR267 pKa = 11.84VLGTNMEE274 pKa = 4.42ANGAGQLFAYY284 pKa = 9.7AVTVGIHH291 pKa = 5.33SVVTVVGMTDD301 pKa = 2.73EE302 pKa = 4.44GAYY305 pKa = 9.32MRR307 pKa = 11.84DD308 pKa = 3.47VFRR311 pKa = 11.84STSFAPSYY319 pKa = 11.02GGISTVLPEE328 pKa = 3.73WHH330 pKa = 6.7GFPRR334 pKa = 11.84PADD337 pKa = 3.38SSLAGWVGLVDD348 pKa = 5.48SIALGTAASVAIADD362 pKa = 3.92PCVAIADD369 pKa = 4.31RR370 pKa = 11.84YY371 pKa = 10.67YY372 pKa = 9.48PTTLTGRR379 pKa = 11.84YY380 pKa = 8.77EE381 pKa = 4.16YY382 pKa = 10.94GGEE385 pKa = 3.88PGMGYY390 pKa = 10.38VGDD393 pKa = 4.26EE394 pKa = 3.66NDD396 pKa = 3.41ARR398 pKa = 11.84IISGKK403 pKa = 9.96IMQLAPSFTRR413 pKa = 11.84NYY415 pKa = 9.92FKK417 pKa = 11.13VLARR421 pKa = 11.84LFSVTPHH428 pKa = 6.34EE429 pKa = 4.39NSSVGLTHH437 pKa = 7.13MIHH440 pKa = 6.61AFSSEE445 pKa = 3.89TLRR448 pKa = 11.84DD449 pKa = 3.69SRR451 pKa = 11.84HH452 pKa = 5.41LQHH455 pKa = 6.99PSVSPFYY462 pKa = 9.42WVEE465 pKa = 3.5PTGIITFDD473 pKa = 3.36TSDD476 pKa = 3.62FTATAAGFGTLATPSQPGTIPMFEE500 pKa = 4.07RR501 pKa = 11.84AEE503 pKa = 4.14VTQNVGDD510 pKa = 3.88VSDD513 pKa = 4.64VLVAWRR519 pKa = 11.84SARR522 pKa = 11.84TSGLAIHH529 pKa = 6.92LNNHH533 pKa = 5.15VEE535 pKa = 4.09AGMEE539 pKa = 4.29NIRR542 pKa = 11.84ITGCDD547 pKa = 3.59PNSWAQLGGASQSVSNRR564 pKa = 11.84VTAGNDD570 pKa = 2.93LASYY574 pKa = 8.2MWARR578 pKa = 11.84SDD580 pKa = 3.56VGVPAPGEE588 pKa = 3.73AMYY591 pKa = 10.69LGEE594 pKa = 5.39AISLLVRR601 pKa = 11.84HH602 pKa = 6.32DD603 pKa = 3.96TLDD606 pKa = 3.54GNTWANTPNHH616 pKa = 5.92VPRR619 pKa = 11.84DD620 pKa = 3.71VEE622 pKa = 4.07FGNNVTIRR630 pKa = 11.84VGLLAPYY637 pKa = 9.96GVGPIGPNRR646 pKa = 11.84KK647 pKa = 9.44ANRR650 pKa = 11.84AFTAALRR657 pKa = 11.84ALDD660 pKa = 3.75AARR663 pKa = 11.84EE664 pKa = 4.22SGMIRR669 pKa = 11.84APKK672 pKa = 10.1LCGTIKK678 pKa = 10.29LTDD681 pKa = 3.37QAFGRR686 pKa = 11.84QAAVPVAIAPPSPKK700 pKa = 10.04AANRR704 pKa = 11.84PVTTVSQPPQSDD716 pKa = 3.25ARR718 pKa = 11.84PVEE721 pKa = 3.98PQNYY725 pKa = 9.03AEE727 pKa = 4.9SPTAHH732 pKa = 7.06VYY734 pKa = 8.2TQGPKK739 pKa = 8.66TAPGPAQSVIASVRR753 pKa = 11.84QMAAEE758 pKa = 4.31EE759 pKa = 4.2SAEE762 pKa = 4.27TASVAAAPQAGAAQQ776 pKa = 3.55
Molecular weight: 81.9 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M9QS71|M9QS71_9VIRU Putative coat protein OS=Ustilaginoidea virens RNA virus 1 OX=1312445 PE=4 SV=1
MM1 pKa = 7.37EE2 pKa = 4.67SAAVRR7 pKa = 11.84ANEE10 pKa = 3.84LGAYY14 pKa = 8.9GHH16 pKa = 7.0ALLRR20 pKa = 11.84FLPPSEE26 pKa = 4.32VCDD29 pKa = 5.17DD30 pKa = 3.28IAMSNTDD37 pKa = 4.1AQWHH41 pKa = 5.17MLGSLGTKK49 pKa = 9.3YY50 pKa = 10.98GYY52 pKa = 9.91ISQVACSLLVCNTPFQVMLHH72 pKa = 5.8EE73 pKa = 4.09RR74 pKa = 11.84HH75 pKa = 5.84IVEE78 pKa = 4.76LLHH81 pKa = 6.62LVKK84 pKa = 10.42PVLTPHH90 pKa = 6.82CEE92 pKa = 3.84YY93 pKa = 11.03DD94 pKa = 3.64VTSPCSACIEE104 pKa = 4.18LDD106 pKa = 3.27SLGRR110 pKa = 11.84VGPGSTVLAFKK121 pKa = 11.06GDD123 pKa = 3.61IAHH126 pKa = 6.47TKK128 pKa = 10.24CNIAFSDD135 pKa = 3.62AWVDD139 pKa = 3.53LCNNSPAIASQVQRR153 pKa = 11.84SGRR156 pKa = 11.84ALHH159 pKa = 6.63GCYY162 pKa = 9.66NDD164 pKa = 3.31QGTAIIMYY172 pKa = 7.82TAALFQHH179 pKa = 6.83LKK181 pKa = 11.05GEE183 pKa = 4.28AFPYY187 pKa = 9.95SYY189 pKa = 10.88HH190 pKa = 6.51FVRR193 pKa = 11.84NPHH196 pKa = 6.1DD197 pKa = 3.88AKK199 pKa = 11.11AFSTILKK206 pKa = 10.69AMGANATPLGSVLTEE221 pKa = 4.01ANTLTGRR228 pKa = 11.84GVNPIDD234 pKa = 3.55VEE236 pKa = 3.94AAGRR240 pKa = 11.84DD241 pKa = 3.09RR242 pKa = 11.84TQYY245 pKa = 11.52DD246 pKa = 3.66KK247 pKa = 10.88IVRR250 pKa = 11.84DD251 pKa = 3.93LFHH254 pKa = 8.02ADD256 pKa = 2.97EE257 pKa = 4.98GKK259 pKa = 10.39LRR261 pKa = 11.84EE262 pKa = 4.6AIRR265 pKa = 11.84TILRR269 pKa = 11.84EE270 pKa = 4.24EE271 pKa = 3.95IGSANVEE278 pKa = 4.29FPSLEE283 pKa = 4.16EE284 pKa = 3.99YY285 pKa = 10.59FSQRR289 pKa = 11.84WLWTVNGSHH298 pKa = 6.85NFTTQRR304 pKa = 11.84FYY306 pKa = 11.36DD307 pKa = 4.33PEE309 pKa = 3.99DD310 pKa = 3.5HH311 pKa = 6.94RR312 pKa = 11.84PPGAARR318 pKa = 11.84MFRR321 pKa = 11.84RR322 pKa = 11.84AYY324 pKa = 10.22AEE326 pKa = 3.96CTSYY330 pKa = 11.62DD331 pKa = 4.84RR332 pKa = 11.84VAAWNGKK339 pKa = 8.08SFFSPSAKK347 pKa = 10.05LEE349 pKa = 4.3HH350 pKa = 6.37GKK352 pKa = 10.03SRR354 pKa = 11.84AIFAGDD360 pKa = 3.12TLTYY364 pKa = 10.54FRR366 pKa = 11.84FQHH369 pKa = 5.61FLKK372 pKa = 10.42HH373 pKa = 7.16VEE375 pKa = 4.18DD376 pKa = 3.96VWTGKK381 pKa = 9.76HH382 pKa = 4.99VLLNPGRR389 pKa = 11.84GGMYY393 pKa = 10.92GMLKK397 pKa = 10.17RR398 pKa = 11.84IRR400 pKa = 11.84GISAGTGVHH409 pKa = 5.67TMMDD413 pKa = 3.51YY414 pKa = 11.28TDD416 pKa = 4.64FNSAHH421 pKa = 6.31TNASMRR427 pKa = 11.84ILFEE431 pKa = 3.88EE432 pKa = 4.2ATSYY436 pKa = 11.01VGYY439 pKa = 10.56DD440 pKa = 3.46PDD442 pKa = 4.36MGALLANSFDD452 pKa = 3.48NSYY455 pKa = 10.07MVTSDD460 pKa = 3.8GPRR463 pKa = 11.84KK464 pKa = 9.44IAGTLMSGHH473 pKa = 7.16RR474 pKa = 11.84ATTFINSVLNRR485 pKa = 11.84AYY487 pKa = 10.87LLVVNPSILRR497 pKa = 11.84LPAVHH502 pKa = 6.63VGDD505 pKa = 4.97DD506 pKa = 4.08VYY508 pKa = 11.48LSPPSLHH515 pKa = 6.8AAAEE519 pKa = 4.1LMDD522 pKa = 4.52DD523 pKa = 3.48VRR525 pKa = 11.84MSGLRR530 pKa = 11.84MNPLKK535 pKa = 10.72QSVGLITGEE544 pKa = 4.1FLRR547 pKa = 11.84MAFGSGSAFGYY558 pKa = 10.32APRR561 pKa = 11.84AIASMISGNWTNSNEE576 pKa = 4.04LTRR579 pKa = 11.84RR580 pKa = 11.84EE581 pKa = 3.98QVEE584 pKa = 4.2NLVTTSWSLTNRR596 pKa = 11.84CRR598 pKa = 11.84NPAAAVLATTALSKK612 pKa = 10.86RR613 pKa = 11.84CGLSKK618 pKa = 10.79CDD620 pKa = 5.53AEE622 pKa = 4.44ALLNGTLALGSGPARR637 pKa = 11.84AGKK640 pKa = 9.22HH641 pKa = 3.52VYY643 pKa = 10.02KK644 pKa = 10.3RR645 pKa = 11.84VEE647 pKa = 3.96IPAPPVSLIPKK658 pKa = 8.91EE659 pKa = 4.19VLSKK663 pKa = 10.83LPSRR667 pKa = 11.84ATDD670 pKa = 3.46DD671 pKa = 3.39YY672 pKa = 11.41LRR674 pKa = 11.84NHH676 pKa = 6.83TSTLEE681 pKa = 3.92RR682 pKa = 11.84TALQLLGTSPKK693 pKa = 9.33VTMLEE698 pKa = 3.7SSYY701 pKa = 11.56AKK703 pKa = 9.38TLRR706 pKa = 11.84PVDD709 pKa = 4.24EE710 pKa = 4.83VPLPMGYY717 pKa = 10.49AKK719 pKa = 10.57EE720 pKa = 4.13WMFGPASVQRR730 pKa = 11.84GVSIDD735 pKa = 3.24EE736 pKa = 4.03CRR738 pKa = 11.84NRR740 pKa = 11.84DD741 pKa = 3.87VIHH744 pKa = 6.67GALSEE749 pKa = 4.29YY750 pKa = 10.29PLIVLFKK757 pKa = 10.18EE758 pKa = 4.29QFTKK762 pKa = 10.7EE763 pKa = 3.62QLAILLRR770 pKa = 11.84MKK772 pKa = 10.66CDD774 pKa = 3.64LYY776 pKa = 11.03TNDD779 pKa = 2.89VRR781 pKa = 11.84VAAFGAEE788 pKa = 3.68ARR790 pKa = 11.84GMVVEE795 pKa = 4.91GWLPRR800 pKa = 11.84ADD802 pKa = 3.47VQHH805 pKa = 6.72ASRR808 pKa = 11.84RR809 pKa = 11.84ATEE812 pKa = 4.24MVLSTSINLYY822 pKa = 10.0FF823 pKa = 4.81
MM1 pKa = 7.37EE2 pKa = 4.67SAAVRR7 pKa = 11.84ANEE10 pKa = 3.84LGAYY14 pKa = 8.9GHH16 pKa = 7.0ALLRR20 pKa = 11.84FLPPSEE26 pKa = 4.32VCDD29 pKa = 5.17DD30 pKa = 3.28IAMSNTDD37 pKa = 4.1AQWHH41 pKa = 5.17MLGSLGTKK49 pKa = 9.3YY50 pKa = 10.98GYY52 pKa = 9.91ISQVACSLLVCNTPFQVMLHH72 pKa = 5.8EE73 pKa = 4.09RR74 pKa = 11.84HH75 pKa = 5.84IVEE78 pKa = 4.76LLHH81 pKa = 6.62LVKK84 pKa = 10.42PVLTPHH90 pKa = 6.82CEE92 pKa = 3.84YY93 pKa = 11.03DD94 pKa = 3.64VTSPCSACIEE104 pKa = 4.18LDD106 pKa = 3.27SLGRR110 pKa = 11.84VGPGSTVLAFKK121 pKa = 11.06GDD123 pKa = 3.61IAHH126 pKa = 6.47TKK128 pKa = 10.24CNIAFSDD135 pKa = 3.62AWVDD139 pKa = 3.53LCNNSPAIASQVQRR153 pKa = 11.84SGRR156 pKa = 11.84ALHH159 pKa = 6.63GCYY162 pKa = 9.66NDD164 pKa = 3.31QGTAIIMYY172 pKa = 7.82TAALFQHH179 pKa = 6.83LKK181 pKa = 11.05GEE183 pKa = 4.28AFPYY187 pKa = 9.95SYY189 pKa = 10.88HH190 pKa = 6.51FVRR193 pKa = 11.84NPHH196 pKa = 6.1DD197 pKa = 3.88AKK199 pKa = 11.11AFSTILKK206 pKa = 10.69AMGANATPLGSVLTEE221 pKa = 4.01ANTLTGRR228 pKa = 11.84GVNPIDD234 pKa = 3.55VEE236 pKa = 3.94AAGRR240 pKa = 11.84DD241 pKa = 3.09RR242 pKa = 11.84TQYY245 pKa = 11.52DD246 pKa = 3.66KK247 pKa = 10.88IVRR250 pKa = 11.84DD251 pKa = 3.93LFHH254 pKa = 8.02ADD256 pKa = 2.97EE257 pKa = 4.98GKK259 pKa = 10.39LRR261 pKa = 11.84EE262 pKa = 4.6AIRR265 pKa = 11.84TILRR269 pKa = 11.84EE270 pKa = 4.24EE271 pKa = 3.95IGSANVEE278 pKa = 4.29FPSLEE283 pKa = 4.16EE284 pKa = 3.99YY285 pKa = 10.59FSQRR289 pKa = 11.84WLWTVNGSHH298 pKa = 6.85NFTTQRR304 pKa = 11.84FYY306 pKa = 11.36DD307 pKa = 4.33PEE309 pKa = 3.99DD310 pKa = 3.5HH311 pKa = 6.94RR312 pKa = 11.84PPGAARR318 pKa = 11.84MFRR321 pKa = 11.84RR322 pKa = 11.84AYY324 pKa = 10.22AEE326 pKa = 3.96CTSYY330 pKa = 11.62DD331 pKa = 4.84RR332 pKa = 11.84VAAWNGKK339 pKa = 8.08SFFSPSAKK347 pKa = 10.05LEE349 pKa = 4.3HH350 pKa = 6.37GKK352 pKa = 10.03SRR354 pKa = 11.84AIFAGDD360 pKa = 3.12TLTYY364 pKa = 10.54FRR366 pKa = 11.84FQHH369 pKa = 5.61FLKK372 pKa = 10.42HH373 pKa = 7.16VEE375 pKa = 4.18DD376 pKa = 3.96VWTGKK381 pKa = 9.76HH382 pKa = 4.99VLLNPGRR389 pKa = 11.84GGMYY393 pKa = 10.92GMLKK397 pKa = 10.17RR398 pKa = 11.84IRR400 pKa = 11.84GISAGTGVHH409 pKa = 5.67TMMDD413 pKa = 3.51YY414 pKa = 11.28TDD416 pKa = 4.64FNSAHH421 pKa = 6.31TNASMRR427 pKa = 11.84ILFEE431 pKa = 3.88EE432 pKa = 4.2ATSYY436 pKa = 11.01VGYY439 pKa = 10.56DD440 pKa = 3.46PDD442 pKa = 4.36MGALLANSFDD452 pKa = 3.48NSYY455 pKa = 10.07MVTSDD460 pKa = 3.8GPRR463 pKa = 11.84KK464 pKa = 9.44IAGTLMSGHH473 pKa = 7.16RR474 pKa = 11.84ATTFINSVLNRR485 pKa = 11.84AYY487 pKa = 10.87LLVVNPSILRR497 pKa = 11.84LPAVHH502 pKa = 6.63VGDD505 pKa = 4.97DD506 pKa = 4.08VYY508 pKa = 11.48LSPPSLHH515 pKa = 6.8AAAEE519 pKa = 4.1LMDD522 pKa = 4.52DD523 pKa = 3.48VRR525 pKa = 11.84MSGLRR530 pKa = 11.84MNPLKK535 pKa = 10.72QSVGLITGEE544 pKa = 4.1FLRR547 pKa = 11.84MAFGSGSAFGYY558 pKa = 10.32APRR561 pKa = 11.84AIASMISGNWTNSNEE576 pKa = 4.04LTRR579 pKa = 11.84RR580 pKa = 11.84EE581 pKa = 3.98QVEE584 pKa = 4.2NLVTTSWSLTNRR596 pKa = 11.84CRR598 pKa = 11.84NPAAAVLATTALSKK612 pKa = 10.86RR613 pKa = 11.84CGLSKK618 pKa = 10.79CDD620 pKa = 5.53AEE622 pKa = 4.44ALLNGTLALGSGPARR637 pKa = 11.84AGKK640 pKa = 9.22HH641 pKa = 3.52VYY643 pKa = 10.02KK644 pKa = 10.3RR645 pKa = 11.84VEE647 pKa = 3.96IPAPPVSLIPKK658 pKa = 8.91EE659 pKa = 4.19VLSKK663 pKa = 10.83LPSRR667 pKa = 11.84ATDD670 pKa = 3.46DD671 pKa = 3.39YY672 pKa = 11.41LRR674 pKa = 11.84NHH676 pKa = 6.83TSTLEE681 pKa = 3.92RR682 pKa = 11.84TALQLLGTSPKK693 pKa = 9.33VTMLEE698 pKa = 3.7SSYY701 pKa = 11.56AKK703 pKa = 9.38TLRR706 pKa = 11.84PVDD709 pKa = 4.24EE710 pKa = 4.83VPLPMGYY717 pKa = 10.49AKK719 pKa = 10.57EE720 pKa = 4.13WMFGPASVQRR730 pKa = 11.84GVSIDD735 pKa = 3.24EE736 pKa = 4.03CRR738 pKa = 11.84NRR740 pKa = 11.84DD741 pKa = 3.87VIHH744 pKa = 6.67GALSEE749 pKa = 4.29YY750 pKa = 10.29PLIVLFKK757 pKa = 10.18EE758 pKa = 4.29QFTKK762 pKa = 10.7EE763 pKa = 3.62QLAILLRR770 pKa = 11.84MKK772 pKa = 10.66CDD774 pKa = 3.64LYY776 pKa = 11.03TNDD779 pKa = 2.89VRR781 pKa = 11.84VAAFGAEE788 pKa = 3.68ARR790 pKa = 11.84GMVVEE795 pKa = 4.91GWLPRR800 pKa = 11.84ADD802 pKa = 3.47VQHH805 pKa = 6.72ASRR808 pKa = 11.84RR809 pKa = 11.84ATEE812 pKa = 4.24MVLSTSINLYY822 pKa = 10.0FF823 pKa = 4.81
Molecular weight: 90.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1599 |
776 |
823 |
799.5 |
86.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.882 ± 1.336 | 1.251 ± 0.425 |
4.378 ± 0.268 | 4.753 ± 0.26 |
3.315 ± 0.426 | 7.942 ± 0.575 |
2.752 ± 0.393 | 4.065 ± 0.131 |
3.064 ± 0.341 | 8.568 ± 1.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.689 ± 0.349 | 4.315 ± 0.046 |
5.754 ± 0.664 | 2.376 ± 0.141 |
5.879 ± 0.598 | 7.567 ± 0.065 |
6.942 ± 0.463 | 7.692 ± 0.75 |
1.188 ± 0.02 | 3.627 ± 0.077 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
