
Ustilaginoidea virens RNA virus 3
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; Victorivirus; unclassified Victorivirus
Average proteome isoelectric point is 7.21
Get precalculated fractions of proteins
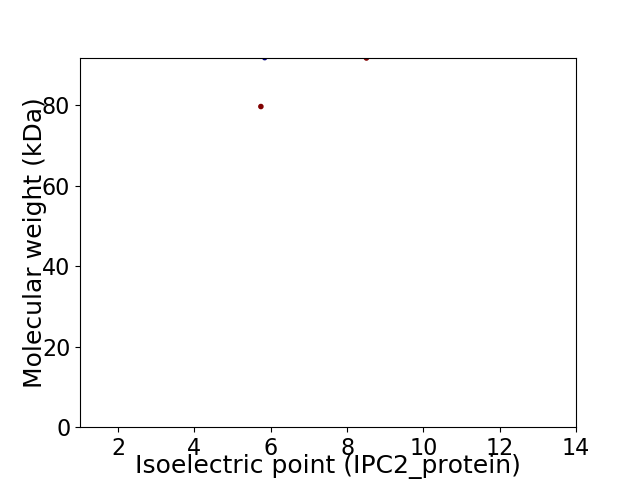
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W5TYK1|W5TYK1_9VIRU Coat protein OS=Ustilaginoidea virens RNA virus 3 OX=1460374 PE=4 SV=1
MM1 pKa = 7.32ATTSRR6 pKa = 11.84FQEE9 pKa = 3.99NNFSYY14 pKa = 10.95LSGGVAGYY22 pKa = 10.47SSSTLVDD29 pKa = 3.44DD30 pKa = 3.75ATYY33 pKa = 10.36RR34 pKa = 11.84RR35 pKa = 11.84YY36 pKa = 9.99RR37 pKa = 11.84AGLTIGTYY45 pKa = 7.53EE46 pKa = 4.26HH47 pKa = 7.1GSYY50 pKa = 9.74TYY52 pKa = 10.18SRR54 pKa = 11.84RR55 pKa = 11.84SIFYY59 pKa = 8.72EE60 pKa = 3.92VGRR63 pKa = 11.84HH64 pKa = 4.73FSGVTKK70 pKa = 10.73ALAYY74 pKa = 10.58NHH76 pKa = 7.22ADD78 pKa = 3.72DD79 pKa = 5.96DD80 pKa = 4.28ALEE83 pKa = 4.64IDD85 pKa = 4.09ASIPINSVEE94 pKa = 3.92AANFEE99 pKa = 3.95GWARR103 pKa = 11.84KK104 pKa = 9.44YY105 pKa = 11.34SNFSPQWHH113 pKa = 6.27MMDD116 pKa = 3.31LCAVVEE122 pKa = 4.21RR123 pKa = 11.84LGKK126 pKa = 10.12AVAAQSVFGGVTTAHH141 pKa = 6.35LRR143 pKa = 11.84GGQPIRR149 pKa = 11.84AVALGTLDD157 pKa = 3.78SPQTASTNSVFIPRR171 pKa = 11.84TVDD174 pKa = 3.12SVGNDD179 pKa = 2.8KK180 pKa = 10.96VFSVLAAAVNGEE192 pKa = 4.09GSTVTTDD199 pKa = 3.68VLRR202 pKa = 11.84LDD204 pKa = 4.07AATNSPIMPEE214 pKa = 4.11CSGHH218 pKa = 6.3GLAKK222 pKa = 10.49ACVEE226 pKa = 4.02ALRR229 pKa = 11.84VLGANFEE236 pKa = 4.22ASGAGDD242 pKa = 4.13LFAYY246 pKa = 10.43ALTRR250 pKa = 11.84GIHH253 pKa = 5.75SGVSVVAHH261 pKa = 6.03TDD263 pKa = 2.89EE264 pKa = 5.22GGWFRR269 pKa = 11.84NVLRR273 pKa = 11.84ACHH276 pKa = 6.02FRR278 pKa = 11.84PPYY281 pKa = 10.61GGINLANRR289 pKa = 11.84DD290 pKa = 4.03YY291 pKa = 11.44PFLPPLAAEE300 pKa = 4.86DD301 pKa = 4.2GGATSAWVDD310 pKa = 4.04SIALKK315 pKa = 9.23TAALVAHH322 pKa = 7.21CDD324 pKa = 3.59PCTHH328 pKa = 6.5ATGGAYY334 pKa = 7.43PTIFVANDD342 pKa = 3.55GAVQPVGTHH351 pKa = 5.81EE352 pKa = 4.14PAGTEE357 pKa = 3.46RR358 pKa = 11.84DD359 pKa = 3.58AASLGGQIAADD370 pKa = 3.58VSRR373 pKa = 11.84FAPLYY378 pKa = 9.33MRR380 pKa = 11.84GLVKK384 pKa = 10.81LFGLRR389 pKa = 11.84ANSGVAEE396 pKa = 4.27SHH398 pKa = 6.65FSTVANVTLSGTKK411 pKa = 9.74DD412 pKa = 2.4RR413 pKa = 11.84HH414 pKa = 5.65LRR416 pKa = 11.84HH417 pKa = 5.83KK418 pKa = 9.21TVAPYY423 pKa = 9.74FFIEE427 pKa = 4.14PTSLIEE433 pKa = 4.18HH434 pKa = 7.1RR435 pKa = 11.84FLGSPAEE442 pKa = 4.06EE443 pKa = 4.25AGFGALTSPGAEE455 pKa = 3.8TEE457 pKa = 4.02LATFEE462 pKa = 4.39RR463 pKa = 11.84VRR465 pKa = 11.84EE466 pKa = 4.01LDD468 pKa = 3.24RR469 pKa = 11.84GKK471 pKa = 9.89HH472 pKa = 6.04ANFCTIAFKK481 pKa = 10.48MRR483 pKa = 11.84TARR486 pKa = 11.84TSGLVAAYY494 pKa = 9.93AGTPAEE500 pKa = 4.03LAGLRR505 pKa = 11.84LYY507 pKa = 10.78QFDD510 pKa = 3.92EE511 pKa = 4.83ASVVLPGDD519 pKa = 4.38LGPTAGTVSAKK530 pKa = 10.03HH531 pKa = 6.07ANADD535 pKa = 3.87PLSSYY540 pKa = 9.4LWKK543 pKa = 10.42RR544 pKa = 11.84GQSPIPAPAEE554 pKa = 3.71FVNIQGSYY562 pKa = 7.49AAKK565 pKa = 8.91YY566 pKa = 9.95HH567 pKa = 5.33VVTWDD572 pKa = 3.55DD573 pKa = 4.54DD574 pKa = 3.99FDD576 pKa = 4.09ATVSDD581 pKa = 5.47LPEE584 pKa = 4.07AWEE587 pKa = 4.52LEE589 pKa = 4.18SHH591 pKa = 5.03PTKK594 pKa = 10.23WRR596 pKa = 11.84VSVPTAIRR604 pKa = 11.84PGATNASDD612 pKa = 3.11NGARR616 pKa = 11.84RR617 pKa = 11.84ARR619 pKa = 11.84CRR621 pKa = 11.84AAIALAQTVIRR632 pKa = 11.84SRR634 pKa = 11.84GLGDD638 pKa = 4.13ANSPCITVSNTPPSWDD654 pKa = 4.37DD655 pKa = 3.16PVQPMARR662 pKa = 11.84KK663 pKa = 9.52EE664 pKa = 4.09DD665 pKa = 4.06TLHH668 pKa = 6.41HH669 pKa = 6.77PGVGTVSSFAEE680 pKa = 4.72GNPPPAVPAALGPPLAPTLTHH701 pKa = 5.89QPLRR705 pKa = 11.84GAPLPRR711 pKa = 11.84HH712 pKa = 5.83GGGVVTGPPPTAAVDD727 pKa = 4.03TPVVAPSAPPSVDD740 pKa = 3.47PRR742 pKa = 11.84PDD744 pKa = 3.72DD745 pKa = 5.03ADD747 pKa = 3.66TATAGPAPRR756 pKa = 11.84II757 pKa = 3.93
MM1 pKa = 7.32ATTSRR6 pKa = 11.84FQEE9 pKa = 3.99NNFSYY14 pKa = 10.95LSGGVAGYY22 pKa = 10.47SSSTLVDD29 pKa = 3.44DD30 pKa = 3.75ATYY33 pKa = 10.36RR34 pKa = 11.84RR35 pKa = 11.84YY36 pKa = 9.99RR37 pKa = 11.84AGLTIGTYY45 pKa = 7.53EE46 pKa = 4.26HH47 pKa = 7.1GSYY50 pKa = 9.74TYY52 pKa = 10.18SRR54 pKa = 11.84RR55 pKa = 11.84SIFYY59 pKa = 8.72EE60 pKa = 3.92VGRR63 pKa = 11.84HH64 pKa = 4.73FSGVTKK70 pKa = 10.73ALAYY74 pKa = 10.58NHH76 pKa = 7.22ADD78 pKa = 3.72DD79 pKa = 5.96DD80 pKa = 4.28ALEE83 pKa = 4.64IDD85 pKa = 4.09ASIPINSVEE94 pKa = 3.92AANFEE99 pKa = 3.95GWARR103 pKa = 11.84KK104 pKa = 9.44YY105 pKa = 11.34SNFSPQWHH113 pKa = 6.27MMDD116 pKa = 3.31LCAVVEE122 pKa = 4.21RR123 pKa = 11.84LGKK126 pKa = 10.12AVAAQSVFGGVTTAHH141 pKa = 6.35LRR143 pKa = 11.84GGQPIRR149 pKa = 11.84AVALGTLDD157 pKa = 3.78SPQTASTNSVFIPRR171 pKa = 11.84TVDD174 pKa = 3.12SVGNDD179 pKa = 2.8KK180 pKa = 10.96VFSVLAAAVNGEE192 pKa = 4.09GSTVTTDD199 pKa = 3.68VLRR202 pKa = 11.84LDD204 pKa = 4.07AATNSPIMPEE214 pKa = 4.11CSGHH218 pKa = 6.3GLAKK222 pKa = 10.49ACVEE226 pKa = 4.02ALRR229 pKa = 11.84VLGANFEE236 pKa = 4.22ASGAGDD242 pKa = 4.13LFAYY246 pKa = 10.43ALTRR250 pKa = 11.84GIHH253 pKa = 5.75SGVSVVAHH261 pKa = 6.03TDD263 pKa = 2.89EE264 pKa = 5.22GGWFRR269 pKa = 11.84NVLRR273 pKa = 11.84ACHH276 pKa = 6.02FRR278 pKa = 11.84PPYY281 pKa = 10.61GGINLANRR289 pKa = 11.84DD290 pKa = 4.03YY291 pKa = 11.44PFLPPLAAEE300 pKa = 4.86DD301 pKa = 4.2GGATSAWVDD310 pKa = 4.04SIALKK315 pKa = 9.23TAALVAHH322 pKa = 7.21CDD324 pKa = 3.59PCTHH328 pKa = 6.5ATGGAYY334 pKa = 7.43PTIFVANDD342 pKa = 3.55GAVQPVGTHH351 pKa = 5.81EE352 pKa = 4.14PAGTEE357 pKa = 3.46RR358 pKa = 11.84DD359 pKa = 3.58AASLGGQIAADD370 pKa = 3.58VSRR373 pKa = 11.84FAPLYY378 pKa = 9.33MRR380 pKa = 11.84GLVKK384 pKa = 10.81LFGLRR389 pKa = 11.84ANSGVAEE396 pKa = 4.27SHH398 pKa = 6.65FSTVANVTLSGTKK411 pKa = 9.74DD412 pKa = 2.4RR413 pKa = 11.84HH414 pKa = 5.65LRR416 pKa = 11.84HH417 pKa = 5.83KK418 pKa = 9.21TVAPYY423 pKa = 9.74FFIEE427 pKa = 4.14PTSLIEE433 pKa = 4.18HH434 pKa = 7.1RR435 pKa = 11.84FLGSPAEE442 pKa = 4.06EE443 pKa = 4.25AGFGALTSPGAEE455 pKa = 3.8TEE457 pKa = 4.02LATFEE462 pKa = 4.39RR463 pKa = 11.84VRR465 pKa = 11.84EE466 pKa = 4.01LDD468 pKa = 3.24RR469 pKa = 11.84GKK471 pKa = 9.89HH472 pKa = 6.04ANFCTIAFKK481 pKa = 10.48MRR483 pKa = 11.84TARR486 pKa = 11.84TSGLVAAYY494 pKa = 9.93AGTPAEE500 pKa = 4.03LAGLRR505 pKa = 11.84LYY507 pKa = 10.78QFDD510 pKa = 3.92EE511 pKa = 4.83ASVVLPGDD519 pKa = 4.38LGPTAGTVSAKK530 pKa = 10.03HH531 pKa = 6.07ANADD535 pKa = 3.87PLSSYY540 pKa = 9.4LWKK543 pKa = 10.42RR544 pKa = 11.84GQSPIPAPAEE554 pKa = 3.71FVNIQGSYY562 pKa = 7.49AAKK565 pKa = 8.91YY566 pKa = 9.95HH567 pKa = 5.33VVTWDD572 pKa = 3.55DD573 pKa = 4.54DD574 pKa = 3.99FDD576 pKa = 4.09ATVSDD581 pKa = 5.47LPEE584 pKa = 4.07AWEE587 pKa = 4.52LEE589 pKa = 4.18SHH591 pKa = 5.03PTKK594 pKa = 10.23WRR596 pKa = 11.84VSVPTAIRR604 pKa = 11.84PGATNASDD612 pKa = 3.11NGARR616 pKa = 11.84RR617 pKa = 11.84ARR619 pKa = 11.84CRR621 pKa = 11.84AAIALAQTVIRR632 pKa = 11.84SRR634 pKa = 11.84GLGDD638 pKa = 4.13ANSPCITVSNTPPSWDD654 pKa = 4.37DD655 pKa = 3.16PVQPMARR662 pKa = 11.84KK663 pKa = 9.52EE664 pKa = 4.09DD665 pKa = 4.06TLHH668 pKa = 6.41HH669 pKa = 6.77PGVGTVSSFAEE680 pKa = 4.72GNPPPAVPAALGPPLAPTLTHH701 pKa = 5.89QPLRR705 pKa = 11.84GAPLPRR711 pKa = 11.84HH712 pKa = 5.83GGGVVTGPPPTAAVDD727 pKa = 4.03TPVVAPSAPPSVDD740 pKa = 3.47PRR742 pKa = 11.84PDD744 pKa = 3.72DD745 pKa = 5.03ADD747 pKa = 3.66TATAGPAPRR756 pKa = 11.84II757 pKa = 3.93
Molecular weight: 79.71 kDa
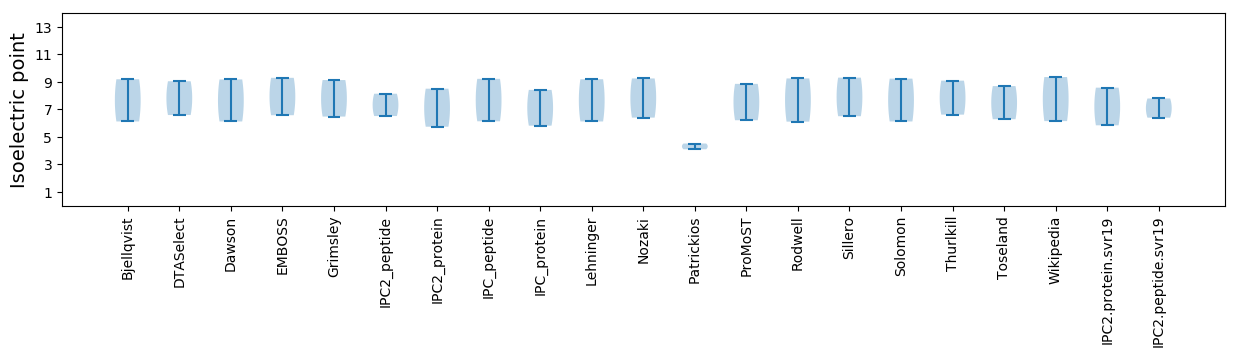
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W5TYK1|W5TYK1_9VIRU Coat protein OS=Ustilaginoidea virens RNA virus 3 OX=1460374 PE=4 SV=1
MM1 pKa = 7.73GDD3 pKa = 4.07DD4 pKa = 4.75VITDD8 pKa = 3.88RR9 pKa = 11.84ANKK12 pKa = 9.88HH13 pKa = 5.53GLLGQYY19 pKa = 9.42LVRR22 pKa = 11.84WLRR25 pKa = 11.84PYY27 pKa = 9.09EE28 pKa = 4.33TGTFTALPLDD38 pKa = 3.78EE39 pKa = 4.26QMSYY43 pKa = 10.06IYY45 pKa = 10.5RR46 pKa = 11.84PFWGSTRR53 pKa = 11.84PPALARR59 pKa = 11.84AALSYY64 pKa = 10.73LGADD68 pKa = 3.37MPIQVVLSDD77 pKa = 4.16SDD79 pKa = 3.73FKK81 pKa = 11.39HH82 pKa = 6.19ILTYY86 pKa = 10.95LPTSPDD92 pKa = 2.98LPATVDD98 pKa = 3.35VPAWVFTKK106 pKa = 10.57AGTMQHH112 pKa = 6.02FRR114 pKa = 11.84PKK116 pKa = 10.06AHH118 pKa = 6.82AKK120 pKa = 10.21AVTKK124 pKa = 10.91ANIFLDD130 pKa = 3.71EE131 pKa = 4.15VLRR134 pKa = 11.84DD135 pKa = 3.89ARR137 pKa = 11.84RR138 pKa = 11.84LDD140 pKa = 3.57ANFVMAAWPYY150 pKa = 11.22LDD152 pKa = 5.15DD153 pKa = 4.65LRR155 pKa = 11.84SNGITHH161 pKa = 6.98DD162 pKa = 3.52QAVAFVLYY170 pKa = 10.41AWALKK175 pKa = 10.4GKK177 pKa = 7.9TVDD180 pKa = 3.37NLRR183 pKa = 11.84WAHH186 pKa = 5.99FCCSQRR192 pKa = 11.84KK193 pKa = 6.41EE194 pKa = 4.08AKK196 pKa = 9.53EE197 pKa = 3.65VSNFLKK203 pKa = 10.78AVGGNAHH210 pKa = 7.29PLGAMMVEE218 pKa = 4.34TDD220 pKa = 3.41TLAGRR225 pKa = 11.84GTGEE229 pKa = 3.98STLLEE234 pKa = 4.38GAIEE238 pKa = 4.33RR239 pKa = 11.84CDD241 pKa = 3.61LAALRR246 pKa = 11.84ASKK249 pKa = 10.37LAEE252 pKa = 3.92FDD254 pKa = 3.87EE255 pKa = 4.77NSLRR259 pKa = 11.84RR260 pKa = 11.84AIRR263 pKa = 11.84RR264 pKa = 11.84ILVSEE269 pKa = 4.46ISRR272 pKa = 11.84NDD274 pKa = 3.09TGGYY278 pKa = 9.78HH279 pKa = 7.04LAFPALSDD287 pKa = 3.13HH288 pKa = 6.48WASRR292 pKa = 11.84WRR294 pKa = 11.84WAVNGSHH301 pKa = 6.02SAEE304 pKa = 4.08VDD306 pKa = 3.67RR307 pKa = 11.84QCGFTPPPLPGGRR320 pKa = 11.84KK321 pKa = 5.08YY322 pKa = 10.38HH323 pKa = 5.71RR324 pKa = 11.84RR325 pKa = 11.84AWLEE329 pKa = 3.62TRR331 pKa = 11.84ADD333 pKa = 3.8DD334 pKa = 5.68PRR336 pKa = 11.84PGWDD340 pKa = 2.71GTTYY344 pKa = 10.87VSASDD349 pKa = 3.53KK350 pKa = 11.12LEE352 pKa = 4.23HH353 pKa = 6.47GKK355 pKa = 8.01TRR357 pKa = 11.84TILACDD363 pKa = 3.42TRR365 pKa = 11.84SYY367 pKa = 11.33LAFEE371 pKa = 4.57HH372 pKa = 7.05LMGTVEE378 pKa = 4.2KK379 pKa = 9.57AWRR382 pKa = 11.84GTRR385 pKa = 11.84VILNPGKK392 pKa = 10.44GGHH395 pKa = 6.45IGMANRR401 pKa = 11.84VEE403 pKa = 4.29RR404 pKa = 11.84NRR406 pKa = 11.84NRR408 pKa = 11.84SGVSMMLDD416 pKa = 3.08YY417 pKa = 11.58DD418 pKa = 4.48DD419 pKa = 5.94FNSHH423 pKa = 6.72HH424 pKa = 7.15SNEE427 pKa = 3.88AMKK430 pKa = 10.54ILVEE434 pKa = 4.36EE435 pKa = 4.69TCQLTGYY442 pKa = 8.8PAEE445 pKa = 4.32LAAPLIASFDD455 pKa = 3.46KK456 pKa = 10.82QRR458 pKa = 11.84IYY460 pKa = 11.2VAGKK464 pKa = 8.55YY465 pKa = 10.07VGVSRR470 pKa = 11.84GTLMSGHH477 pKa = 6.99RR478 pKa = 11.84CTTYY482 pKa = 10.57INSVLNMAYY491 pKa = 10.86LMVVLGDD498 pKa = 3.92DD499 pKa = 4.11FVMEE503 pKa = 4.64RR504 pKa = 11.84PTLHH508 pKa = 6.81VGDD511 pKa = 4.79DD512 pKa = 3.53VFFGVRR518 pKa = 11.84SYY520 pKa = 11.8SEE522 pKa = 3.71ACYY525 pKa = 10.04VARR528 pKa = 11.84SVLASRR534 pKa = 11.84LRR536 pKa = 11.84MNRR539 pKa = 11.84SKK541 pKa = 11.04QSVGHH546 pKa = 5.87VATEE550 pKa = 3.89FLRR553 pKa = 11.84VSSRR557 pKa = 11.84ARR559 pKa = 11.84DD560 pKa = 3.38SYY562 pKa = 11.81GYY564 pKa = 9.97LCRR567 pKa = 11.84AISSCVSGNWVSDD580 pKa = 3.52KK581 pKa = 11.31LLDD584 pKa = 4.33PYY586 pKa = 10.9EE587 pKa = 4.73ALNTMLGSARR597 pKa = 11.84TLMNRR602 pKa = 11.84AISHH606 pKa = 6.85DD607 pKa = 4.48LPLLLASAIKK617 pKa = 9.49RR618 pKa = 11.84TVRR621 pKa = 11.84ADD623 pKa = 3.43GLNDD627 pKa = 3.75RR628 pKa = 11.84LLTEE632 pKa = 4.7LLLGSVAVNNGPQFSSSGTHH652 pKa = 6.08RR653 pKa = 11.84IVWVQPQSRR662 pKa = 11.84LLDD665 pKa = 4.18PLDD668 pKa = 3.7VSLLPRR674 pKa = 11.84EE675 pKa = 4.35STASYY680 pKa = 10.56LVNCAQPIEE689 pKa = 4.42TQTLARR695 pKa = 11.84VGISPVGTMLASSYY709 pKa = 11.29AKK711 pKa = 10.38SLDD714 pKa = 3.51YY715 pKa = 11.31ARR717 pKa = 11.84TQTFRR722 pKa = 11.84LSFSAVHH729 pKa = 6.64TYY731 pKa = 10.41RR732 pKa = 11.84AVGSTTAEE740 pKa = 4.02LALASPKK747 pKa = 10.01PRR749 pKa = 11.84GILNQYY755 pKa = 9.19PLLVLVKK762 pKa = 10.3HH763 pKa = 6.36RR764 pKa = 11.84LPEE767 pKa = 3.99NVLRR771 pKa = 11.84EE772 pKa = 4.12VVATVGGNGHH782 pKa = 6.61AEE784 pKa = 4.14RR785 pKa = 11.84IVVEE789 pKa = 3.62AWGEE793 pKa = 3.92YY794 pKa = 9.04RR795 pKa = 11.84HH796 pKa = 6.08GCIINSVLSFSDD808 pKa = 3.88ASALGKK814 pKa = 9.85RR815 pKa = 11.84VKK817 pKa = 10.83ASVLTSGRR825 pKa = 11.84HH826 pKa = 5.33CYY828 pKa = 8.68VV829 pKa = 2.93
MM1 pKa = 7.73GDD3 pKa = 4.07DD4 pKa = 4.75VITDD8 pKa = 3.88RR9 pKa = 11.84ANKK12 pKa = 9.88HH13 pKa = 5.53GLLGQYY19 pKa = 9.42LVRR22 pKa = 11.84WLRR25 pKa = 11.84PYY27 pKa = 9.09EE28 pKa = 4.33TGTFTALPLDD38 pKa = 3.78EE39 pKa = 4.26QMSYY43 pKa = 10.06IYY45 pKa = 10.5RR46 pKa = 11.84PFWGSTRR53 pKa = 11.84PPALARR59 pKa = 11.84AALSYY64 pKa = 10.73LGADD68 pKa = 3.37MPIQVVLSDD77 pKa = 4.16SDD79 pKa = 3.73FKK81 pKa = 11.39HH82 pKa = 6.19ILTYY86 pKa = 10.95LPTSPDD92 pKa = 2.98LPATVDD98 pKa = 3.35VPAWVFTKK106 pKa = 10.57AGTMQHH112 pKa = 6.02FRR114 pKa = 11.84PKK116 pKa = 10.06AHH118 pKa = 6.82AKK120 pKa = 10.21AVTKK124 pKa = 10.91ANIFLDD130 pKa = 3.71EE131 pKa = 4.15VLRR134 pKa = 11.84DD135 pKa = 3.89ARR137 pKa = 11.84RR138 pKa = 11.84LDD140 pKa = 3.57ANFVMAAWPYY150 pKa = 11.22LDD152 pKa = 5.15DD153 pKa = 4.65LRR155 pKa = 11.84SNGITHH161 pKa = 6.98DD162 pKa = 3.52QAVAFVLYY170 pKa = 10.41AWALKK175 pKa = 10.4GKK177 pKa = 7.9TVDD180 pKa = 3.37NLRR183 pKa = 11.84WAHH186 pKa = 5.99FCCSQRR192 pKa = 11.84KK193 pKa = 6.41EE194 pKa = 4.08AKK196 pKa = 9.53EE197 pKa = 3.65VSNFLKK203 pKa = 10.78AVGGNAHH210 pKa = 7.29PLGAMMVEE218 pKa = 4.34TDD220 pKa = 3.41TLAGRR225 pKa = 11.84GTGEE229 pKa = 3.98STLLEE234 pKa = 4.38GAIEE238 pKa = 4.33RR239 pKa = 11.84CDD241 pKa = 3.61LAALRR246 pKa = 11.84ASKK249 pKa = 10.37LAEE252 pKa = 3.92FDD254 pKa = 3.87EE255 pKa = 4.77NSLRR259 pKa = 11.84RR260 pKa = 11.84AIRR263 pKa = 11.84RR264 pKa = 11.84ILVSEE269 pKa = 4.46ISRR272 pKa = 11.84NDD274 pKa = 3.09TGGYY278 pKa = 9.78HH279 pKa = 7.04LAFPALSDD287 pKa = 3.13HH288 pKa = 6.48WASRR292 pKa = 11.84WRR294 pKa = 11.84WAVNGSHH301 pKa = 6.02SAEE304 pKa = 4.08VDD306 pKa = 3.67RR307 pKa = 11.84QCGFTPPPLPGGRR320 pKa = 11.84KK321 pKa = 5.08YY322 pKa = 10.38HH323 pKa = 5.71RR324 pKa = 11.84RR325 pKa = 11.84AWLEE329 pKa = 3.62TRR331 pKa = 11.84ADD333 pKa = 3.8DD334 pKa = 5.68PRR336 pKa = 11.84PGWDD340 pKa = 2.71GTTYY344 pKa = 10.87VSASDD349 pKa = 3.53KK350 pKa = 11.12LEE352 pKa = 4.23HH353 pKa = 6.47GKK355 pKa = 8.01TRR357 pKa = 11.84TILACDD363 pKa = 3.42TRR365 pKa = 11.84SYY367 pKa = 11.33LAFEE371 pKa = 4.57HH372 pKa = 7.05LMGTVEE378 pKa = 4.2KK379 pKa = 9.57AWRR382 pKa = 11.84GTRR385 pKa = 11.84VILNPGKK392 pKa = 10.44GGHH395 pKa = 6.45IGMANRR401 pKa = 11.84VEE403 pKa = 4.29RR404 pKa = 11.84NRR406 pKa = 11.84NRR408 pKa = 11.84SGVSMMLDD416 pKa = 3.08YY417 pKa = 11.58DD418 pKa = 4.48DD419 pKa = 5.94FNSHH423 pKa = 6.72HH424 pKa = 7.15SNEE427 pKa = 3.88AMKK430 pKa = 10.54ILVEE434 pKa = 4.36EE435 pKa = 4.69TCQLTGYY442 pKa = 8.8PAEE445 pKa = 4.32LAAPLIASFDD455 pKa = 3.46KK456 pKa = 10.82QRR458 pKa = 11.84IYY460 pKa = 11.2VAGKK464 pKa = 8.55YY465 pKa = 10.07VGVSRR470 pKa = 11.84GTLMSGHH477 pKa = 6.99RR478 pKa = 11.84CTTYY482 pKa = 10.57INSVLNMAYY491 pKa = 10.86LMVVLGDD498 pKa = 3.92DD499 pKa = 4.11FVMEE503 pKa = 4.64RR504 pKa = 11.84PTLHH508 pKa = 6.81VGDD511 pKa = 4.79DD512 pKa = 3.53VFFGVRR518 pKa = 11.84SYY520 pKa = 11.8SEE522 pKa = 3.71ACYY525 pKa = 10.04VARR528 pKa = 11.84SVLASRR534 pKa = 11.84LRR536 pKa = 11.84MNRR539 pKa = 11.84SKK541 pKa = 11.04QSVGHH546 pKa = 5.87VATEE550 pKa = 3.89FLRR553 pKa = 11.84VSSRR557 pKa = 11.84ARR559 pKa = 11.84DD560 pKa = 3.38SYY562 pKa = 11.81GYY564 pKa = 9.97LCRR567 pKa = 11.84AISSCVSGNWVSDD580 pKa = 3.52KK581 pKa = 11.31LLDD584 pKa = 4.33PYY586 pKa = 10.9EE587 pKa = 4.73ALNTMLGSARR597 pKa = 11.84TLMNRR602 pKa = 11.84AISHH606 pKa = 6.85DD607 pKa = 4.48LPLLLASAIKK617 pKa = 9.49RR618 pKa = 11.84TVRR621 pKa = 11.84ADD623 pKa = 3.43GLNDD627 pKa = 3.75RR628 pKa = 11.84LLTEE632 pKa = 4.7LLLGSVAVNNGPQFSSSGTHH652 pKa = 6.08RR653 pKa = 11.84IVWVQPQSRR662 pKa = 11.84LLDD665 pKa = 4.18PLDD668 pKa = 3.7VSLLPRR674 pKa = 11.84EE675 pKa = 4.35STASYY680 pKa = 10.56LVNCAQPIEE689 pKa = 4.42TQTLARR695 pKa = 11.84VGISPVGTMLASSYY709 pKa = 11.29AKK711 pKa = 10.38SLDD714 pKa = 3.51YY715 pKa = 11.31ARR717 pKa = 11.84TQTFRR722 pKa = 11.84LSFSAVHH729 pKa = 6.64TYY731 pKa = 10.41RR732 pKa = 11.84AVGSTTAEE740 pKa = 4.02LALASPKK747 pKa = 10.01PRR749 pKa = 11.84GILNQYY755 pKa = 9.19PLLVLVKK762 pKa = 10.3HH763 pKa = 6.36RR764 pKa = 11.84LPEE767 pKa = 3.99NVLRR771 pKa = 11.84EE772 pKa = 4.12VVATVGGNGHH782 pKa = 6.61AEE784 pKa = 4.14RR785 pKa = 11.84IVVEE789 pKa = 3.62AWGEE793 pKa = 3.92YY794 pKa = 9.04RR795 pKa = 11.84HH796 pKa = 6.08GCIINSVLSFSDD808 pKa = 3.88ASALGKK814 pKa = 9.85RR815 pKa = 11.84VKK817 pKa = 10.83ASVLTSGRR825 pKa = 11.84HH826 pKa = 5.33CYY828 pKa = 8.68VV829 pKa = 2.93
Molecular weight: 91.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1586 |
757 |
829 |
793.0 |
85.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.98 ± 1.26 | 1.387 ± 0.116 |
5.422 ± 0.004 | 4.224 ± 0.002 |
3.342 ± 0.286 | 8.071 ± 0.611 |
3.153 ± 0.01 | 3.153 ± 0.067 |
2.774 ± 0.386 | 9.079 ± 1.138 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.702 ± 0.455 | 3.468 ± 0.097 |
5.99 ± 1.055 | 1.892 ± 0.102 |
7.251 ± 0.687 | 7.755 ± 0.209 |
6.81 ± 0.498 | 7.818 ± 0.014 |
1.513 ± 0.19 | 3.216 ± 0.258 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
