
Sewage-associated circular DNA virus-30
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 7.53
Get precalculated fractions of proteins
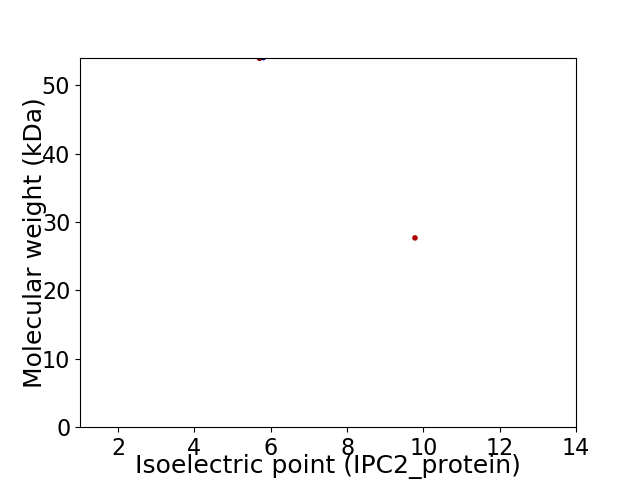
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4UI52|A0A0B4UI52_9VIRU Capsid protein OS=Sewage-associated circular DNA virus-30 OX=1592097 PE=4 SV=1
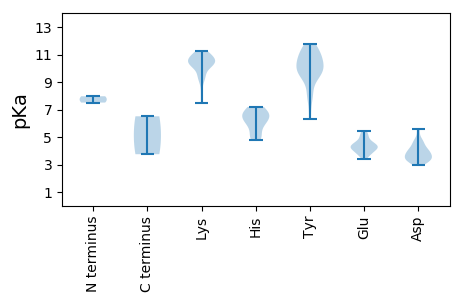
MM1 pKa = 7.51SNFRR5 pKa = 11.84FHH7 pKa = 7.08SKK9 pKa = 10.52SIFLTFPQCDD19 pKa = 3.56YY20 pKa = 10.7PLKK23 pKa = 10.51NFKK26 pKa = 10.91DD27 pKa = 3.52NIEE30 pKa = 4.44AYY32 pKa = 10.55FGDD35 pKa = 3.98NLEE38 pKa = 4.59KK39 pKa = 10.88GVVSQEE45 pKa = 3.42NHH47 pKa = 4.99QDD49 pKa = 3.66GNKK52 pKa = 9.34HH53 pKa = 4.81LHH55 pKa = 6.13AAICLHH61 pKa = 5.15QQVTSRR67 pKa = 11.84DD68 pKa = 3.31PKK70 pKa = 11.13LFDD73 pKa = 4.76KK74 pKa = 11.15LVDD77 pKa = 3.81PAKK80 pKa = 10.34HH81 pKa = 5.81PNIAGRR87 pKa = 11.84FTGGMLKK94 pKa = 10.68AFDD97 pKa = 3.88YY98 pKa = 11.29VMKK101 pKa = 10.37EE102 pKa = 4.15GNFLPLNEE110 pKa = 4.74KK111 pKa = 10.77SFDD114 pKa = 3.56LKK116 pKa = 11.18EE117 pKa = 3.8FLKK120 pKa = 10.9LSKK123 pKa = 10.2EE124 pKa = 4.3KK125 pKa = 10.7KK126 pKa = 9.28NSRR129 pKa = 11.84ASLIVKK135 pKa = 9.46EE136 pKa = 4.41FDD138 pKa = 3.29EE139 pKa = 5.23LPPEE143 pKa = 5.01DD144 pKa = 4.29VMEE147 pKa = 4.36NNKK150 pKa = 10.42DD151 pKa = 3.37FMLLHH156 pKa = 6.7GKK158 pKa = 8.9QMQAYY163 pKa = 8.18VDD165 pKa = 3.31WRR167 pKa = 11.84EE168 pKa = 3.66EE169 pKa = 3.78RR170 pKa = 11.84EE171 pKa = 4.06RR172 pKa = 11.84RR173 pKa = 11.84SKK175 pKa = 10.48FARR178 pKa = 11.84AQAQKK183 pKa = 10.63VFVVPAPGYY192 pKa = 9.6FNAWNNEE199 pKa = 3.54IASWLTTNLRR209 pKa = 11.84QKK211 pKa = 10.21RR212 pKa = 11.84KK213 pKa = 9.43HH214 pKa = 5.94RR215 pKa = 11.84QTQLWIQGPPGIGKK229 pKa = 8.2TSMMMMLEE237 pKa = 4.1EE238 pKa = 5.11LYY240 pKa = 11.1SLTIYY245 pKa = 9.94HH246 pKa = 6.47WPKK249 pKa = 10.91DD250 pKa = 3.85EE251 pKa = 4.51RR252 pKa = 11.84WWDD255 pKa = 3.64LYY257 pKa = 11.74GDD259 pKa = 3.9GQYY262 pKa = 11.47DD263 pKa = 4.15VILLDD268 pKa = 3.62EE269 pKa = 4.71FRR271 pKa = 11.84SQKK274 pKa = 10.15MITEE278 pKa = 4.71LNPILSGDD286 pKa = 4.16PISLSRR292 pKa = 11.84RR293 pKa = 11.84GMAPIVKK300 pKa = 9.38RR301 pKa = 11.84DD302 pKa = 3.83NLPVIIMSNYY312 pKa = 9.7LPEE315 pKa = 4.34EE316 pKa = 4.34CFPTANEE323 pKa = 4.1HH324 pKa = 6.66GKK326 pKa = 10.41LEE328 pKa = 4.25PLLDD332 pKa = 4.64RR333 pKa = 11.84IKK335 pKa = 10.78VVKK338 pKa = 10.56CEE340 pKa = 3.81GPIRR344 pKa = 11.84IVEE347 pKa = 4.17GAPPAEE353 pKa = 4.66CSTPFLFDD361 pKa = 4.32DD362 pKa = 4.67QPPPPPGSQFVLTNDD377 pKa = 3.16YY378 pKa = 11.48SEE380 pKa = 5.47IFPQTPPEE388 pKa = 4.28YY389 pKa = 9.84EE390 pKa = 4.29TIDD393 pKa = 5.45DD394 pKa = 5.58DD395 pKa = 4.7DD396 pKa = 4.45LQPGRR401 pKa = 11.84VCNARR406 pKa = 11.84PQQSTYY412 pKa = 11.22LHH414 pKa = 6.88IFPEE418 pKa = 3.91NRR420 pKa = 11.84EE421 pKa = 3.77AYY423 pKa = 9.72ARR425 pKa = 11.84YY426 pKa = 9.38HH427 pKa = 6.46GDD429 pKa = 3.35AAVVPYY435 pKa = 9.81TSADD439 pKa = 3.3FDD441 pKa = 4.82RR442 pKa = 11.84NNDD445 pKa = 3.0PFYY448 pKa = 10.56FDD450 pKa = 4.06KK451 pKa = 10.73ISRR454 pKa = 11.84QSRR457 pKa = 11.84AAMLAARR464 pKa = 11.84NII466 pKa = 3.75
MM1 pKa = 7.51SNFRR5 pKa = 11.84FHH7 pKa = 7.08SKK9 pKa = 10.52SIFLTFPQCDD19 pKa = 3.56YY20 pKa = 10.7PLKK23 pKa = 10.51NFKK26 pKa = 10.91DD27 pKa = 3.52NIEE30 pKa = 4.44AYY32 pKa = 10.55FGDD35 pKa = 3.98NLEE38 pKa = 4.59KK39 pKa = 10.88GVVSQEE45 pKa = 3.42NHH47 pKa = 4.99QDD49 pKa = 3.66GNKK52 pKa = 9.34HH53 pKa = 4.81LHH55 pKa = 6.13AAICLHH61 pKa = 5.15QQVTSRR67 pKa = 11.84DD68 pKa = 3.31PKK70 pKa = 11.13LFDD73 pKa = 4.76KK74 pKa = 11.15LVDD77 pKa = 3.81PAKK80 pKa = 10.34HH81 pKa = 5.81PNIAGRR87 pKa = 11.84FTGGMLKK94 pKa = 10.68AFDD97 pKa = 3.88YY98 pKa = 11.29VMKK101 pKa = 10.37EE102 pKa = 4.15GNFLPLNEE110 pKa = 4.74KK111 pKa = 10.77SFDD114 pKa = 3.56LKK116 pKa = 11.18EE117 pKa = 3.8FLKK120 pKa = 10.9LSKK123 pKa = 10.2EE124 pKa = 4.3KK125 pKa = 10.7KK126 pKa = 9.28NSRR129 pKa = 11.84ASLIVKK135 pKa = 9.46EE136 pKa = 4.41FDD138 pKa = 3.29EE139 pKa = 5.23LPPEE143 pKa = 5.01DD144 pKa = 4.29VMEE147 pKa = 4.36NNKK150 pKa = 10.42DD151 pKa = 3.37FMLLHH156 pKa = 6.7GKK158 pKa = 8.9QMQAYY163 pKa = 8.18VDD165 pKa = 3.31WRR167 pKa = 11.84EE168 pKa = 3.66EE169 pKa = 3.78RR170 pKa = 11.84EE171 pKa = 4.06RR172 pKa = 11.84RR173 pKa = 11.84SKK175 pKa = 10.48FARR178 pKa = 11.84AQAQKK183 pKa = 10.63VFVVPAPGYY192 pKa = 9.6FNAWNNEE199 pKa = 3.54IASWLTTNLRR209 pKa = 11.84QKK211 pKa = 10.21RR212 pKa = 11.84KK213 pKa = 9.43HH214 pKa = 5.94RR215 pKa = 11.84QTQLWIQGPPGIGKK229 pKa = 8.2TSMMMMLEE237 pKa = 4.1EE238 pKa = 5.11LYY240 pKa = 11.1SLTIYY245 pKa = 9.94HH246 pKa = 6.47WPKK249 pKa = 10.91DD250 pKa = 3.85EE251 pKa = 4.51RR252 pKa = 11.84WWDD255 pKa = 3.64LYY257 pKa = 11.74GDD259 pKa = 3.9GQYY262 pKa = 11.47DD263 pKa = 4.15VILLDD268 pKa = 3.62EE269 pKa = 4.71FRR271 pKa = 11.84SQKK274 pKa = 10.15MITEE278 pKa = 4.71LNPILSGDD286 pKa = 4.16PISLSRR292 pKa = 11.84RR293 pKa = 11.84GMAPIVKK300 pKa = 9.38RR301 pKa = 11.84DD302 pKa = 3.83NLPVIIMSNYY312 pKa = 9.7LPEE315 pKa = 4.34EE316 pKa = 4.34CFPTANEE323 pKa = 4.1HH324 pKa = 6.66GKK326 pKa = 10.41LEE328 pKa = 4.25PLLDD332 pKa = 4.64RR333 pKa = 11.84IKK335 pKa = 10.78VVKK338 pKa = 10.56CEE340 pKa = 3.81GPIRR344 pKa = 11.84IVEE347 pKa = 4.17GAPPAEE353 pKa = 4.66CSTPFLFDD361 pKa = 4.32DD362 pKa = 4.67QPPPPPGSQFVLTNDD377 pKa = 3.16YY378 pKa = 11.48SEE380 pKa = 5.47IFPQTPPEE388 pKa = 4.28YY389 pKa = 9.84EE390 pKa = 4.29TIDD393 pKa = 5.45DD394 pKa = 5.58DD395 pKa = 4.7DD396 pKa = 4.45LQPGRR401 pKa = 11.84VCNARR406 pKa = 11.84PQQSTYY412 pKa = 11.22LHH414 pKa = 6.88IFPEE418 pKa = 3.91NRR420 pKa = 11.84EE421 pKa = 3.77AYY423 pKa = 9.72ARR425 pKa = 11.84YY426 pKa = 9.38HH427 pKa = 6.46GDD429 pKa = 3.35AAVVPYY435 pKa = 9.81TSADD439 pKa = 3.3FDD441 pKa = 4.82RR442 pKa = 11.84NNDD445 pKa = 3.0PFYY448 pKa = 10.56FDD450 pKa = 4.06KK451 pKa = 10.73ISRR454 pKa = 11.84QSRR457 pKa = 11.84AAMLAARR464 pKa = 11.84NII466 pKa = 3.75
Molecular weight: 53.92 kDa
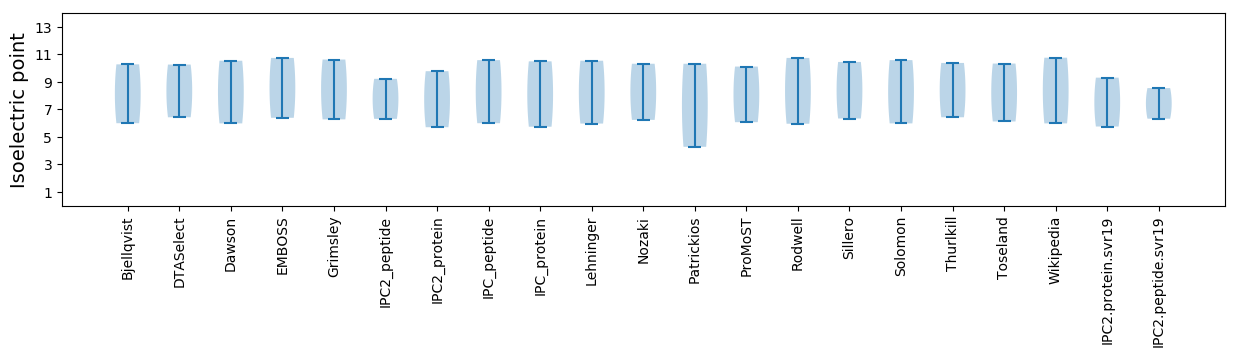
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UI52|A0A0B4UI52_9VIRU Capsid protein OS=Sewage-associated circular DNA virus-30 OX=1592097 PE=4 SV=1
MM1 pKa = 7.97AYY3 pKa = 9.58PRR5 pKa = 11.84GRR7 pKa = 11.84RR8 pKa = 11.84TPLKK12 pKa = 10.33RR13 pKa = 11.84RR14 pKa = 11.84ATYY17 pKa = 10.66GRR19 pKa = 11.84GTRR22 pKa = 11.84TFARR26 pKa = 11.84TKK28 pKa = 10.19FRR30 pKa = 11.84RR31 pKa = 11.84AYY33 pKa = 8.21FNRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84GVGRR42 pKa = 11.84KK43 pKa = 7.48EE44 pKa = 4.02KK45 pKa = 10.37KK46 pKa = 10.08GCDD49 pKa = 3.39VSFATAGIIASTSTNGGIYY68 pKa = 10.02LLNGIQEE75 pKa = 5.07GVGSWNRR82 pKa = 11.84IGRR85 pKa = 11.84YY86 pKa = 6.34MWNKK90 pKa = 10.31SIEE93 pKa = 4.19LDD95 pKa = 3.17LTLRR99 pKa = 11.84YY100 pKa = 7.76TSSYY104 pKa = 10.13GALDD108 pKa = 3.43TDD110 pKa = 3.62ITSGEE115 pKa = 4.04WVRR118 pKa = 11.84AVLVWDD124 pKa = 3.8KK125 pKa = 11.02QPNGGTIPTFDD136 pKa = 4.05TIFGQTSQAGVEE148 pKa = 4.43SASVMDD154 pKa = 4.1HH155 pKa = 6.38LRR157 pKa = 11.84YY158 pKa = 10.85DD159 pKa = 3.03NMFRR163 pKa = 11.84FKK165 pKa = 11.26VLMDD169 pKa = 3.29EE170 pKa = 5.3CINPTLTNAAVSTPINPASASLVNNTYY197 pKa = 10.11FRR199 pKa = 11.84FHH201 pKa = 7.19KK202 pKa = 10.24YY203 pKa = 9.41VKK205 pKa = 9.82LRR207 pKa = 11.84NMPTNFSGTANPMTTANISTGALYY231 pKa = 10.76LILRR235 pKa = 11.84TQSATLTEE243 pKa = 4.48LWGHH247 pKa = 6.51
MM1 pKa = 7.97AYY3 pKa = 9.58PRR5 pKa = 11.84GRR7 pKa = 11.84RR8 pKa = 11.84TPLKK12 pKa = 10.33RR13 pKa = 11.84RR14 pKa = 11.84ATYY17 pKa = 10.66GRR19 pKa = 11.84GTRR22 pKa = 11.84TFARR26 pKa = 11.84TKK28 pKa = 10.19FRR30 pKa = 11.84RR31 pKa = 11.84AYY33 pKa = 8.21FNRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84GVGRR42 pKa = 11.84KK43 pKa = 7.48EE44 pKa = 4.02KK45 pKa = 10.37KK46 pKa = 10.08GCDD49 pKa = 3.39VSFATAGIIASTSTNGGIYY68 pKa = 10.02LLNGIQEE75 pKa = 5.07GVGSWNRR82 pKa = 11.84IGRR85 pKa = 11.84YY86 pKa = 6.34MWNKK90 pKa = 10.31SIEE93 pKa = 4.19LDD95 pKa = 3.17LTLRR99 pKa = 11.84YY100 pKa = 7.76TSSYY104 pKa = 10.13GALDD108 pKa = 3.43TDD110 pKa = 3.62ITSGEE115 pKa = 4.04WVRR118 pKa = 11.84AVLVWDD124 pKa = 3.8KK125 pKa = 11.02QPNGGTIPTFDD136 pKa = 4.05TIFGQTSQAGVEE148 pKa = 4.43SASVMDD154 pKa = 4.1HH155 pKa = 6.38LRR157 pKa = 11.84YY158 pKa = 10.85DD159 pKa = 3.03NMFRR163 pKa = 11.84FKK165 pKa = 11.26VLMDD169 pKa = 3.29EE170 pKa = 5.3CINPTLTNAAVSTPINPASASLVNNTYY197 pKa = 10.11FRR199 pKa = 11.84FHH201 pKa = 7.19KK202 pKa = 10.24YY203 pKa = 9.41VKK205 pKa = 9.82LRR207 pKa = 11.84NMPTNFSGTANPMTTANISTGALYY231 pKa = 10.76LILRR235 pKa = 11.84TQSATLTEE243 pKa = 4.48LWGHH247 pKa = 6.51
Molecular weight: 27.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
713 |
247 |
466 |
356.5 |
40.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.592 ± 0.585 | 1.122 ± 0.166 |
5.891 ± 1.194 | 5.47 ± 1.401 |
5.33 ± 0.466 | 6.171 ± 1.454 |
2.104 ± 0.473 | 5.189 ± 0.039 |
5.75 ± 0.905 | 7.994 ± 0.376 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.945 ± 0.059 | 5.891 ± 0.312 |
6.452 ± 1.493 | 3.787 ± 0.937 |
7.013 ± 1.222 | 5.891 ± 0.527 |
6.171 ± 2.746 | 4.628 ± 0.122 |
1.683 ± 0.181 | 3.927 ± 0.28 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
