
Hortaea werneckii EXF-2000
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; dothideomyceta; Dothideomycetes; Dothideomycetidae; Mycosphaerellales; Teratosphaeriaceae; Hortaea; Hortaea werneckii
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
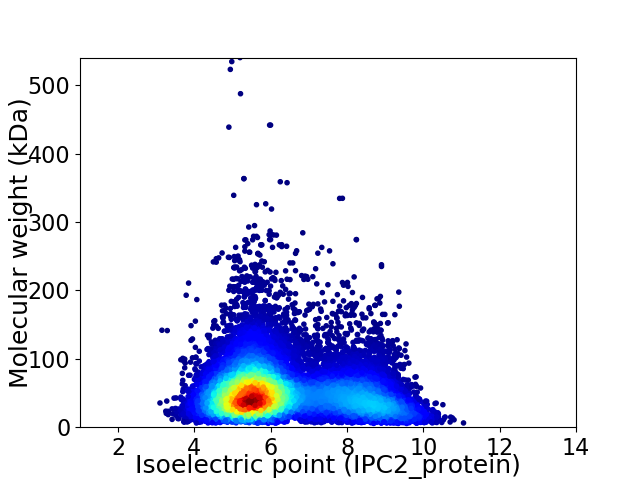
Virtual 2D-PAGE plot for 15464 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Z5T4S0|A0A1Z5T4S0_HORWE Uncharacterized protein OS=Hortaea werneckii EXF-2000 OX=1157616 GN=BTJ68_08764 PE=3 SV=1
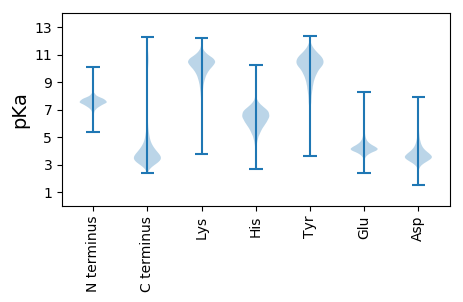
MM1 pKa = 7.36PRR3 pKa = 11.84SHH5 pKa = 6.56GRR7 pKa = 11.84RR8 pKa = 11.84TTAVLLTAAAAAATTVSALPEE29 pKa = 3.58IAITKK34 pKa = 10.04RR35 pKa = 11.84DD36 pKa = 3.7GEE38 pKa = 4.41LKK40 pKa = 10.14ASEE43 pKa = 4.14RR44 pKa = 11.84ARR46 pKa = 11.84GVEE49 pKa = 3.99KK50 pKa = 10.56RR51 pKa = 11.84AADD54 pKa = 3.55GNGTVEE60 pKa = 4.25TNVFDD65 pKa = 3.62VRR67 pKa = 11.84TWSTGGAYY75 pKa = 9.44YY76 pKa = 11.29ANVTVGTPPQPQVVILDD93 pKa = 3.84TGSSDD98 pKa = 4.8LYY100 pKa = 11.05FDD102 pKa = 4.79AADD105 pKa = 4.62APTCSLTGKK114 pKa = 8.35YY115 pKa = 9.41ACKK118 pKa = 10.43GGTFSPDD125 pKa = 2.75NSSTYY130 pKa = 10.34QVVDD134 pKa = 4.01PFPAFDD140 pKa = 3.8TLFGDD145 pKa = 4.74GSSATGEE152 pKa = 4.16YY153 pKa = 11.18GEE155 pKa = 4.52DD156 pKa = 3.16TVGIGDD162 pKa = 3.57VRR164 pKa = 11.84IEE166 pKa = 3.93NVQFGLANSLYY177 pKa = 8.45ITTGYY182 pKa = 11.18AVGLMGLGYY191 pKa = 10.34SYY193 pKa = 11.08IEE195 pKa = 5.15AIDD198 pKa = 4.02DD199 pKa = 4.39TEE201 pKa = 4.28DD202 pKa = 3.6LYY204 pKa = 10.93PNIPEE209 pKa = 4.05VLRR212 pKa = 11.84ATGVINSRR220 pKa = 11.84LYY222 pKa = 10.61SVYY225 pKa = 10.9LNDD228 pKa = 4.98LSDD231 pKa = 3.33ISGTILFGGIDD242 pKa = 3.61PSKK245 pKa = 9.79YY246 pKa = 9.3TGDD249 pKa = 3.53LTTLNILPDD258 pKa = 4.11VISGGISMFLTTVTDD273 pKa = 4.33LSITTDD279 pKa = 3.39GEE281 pKa = 4.35TADD284 pKa = 4.46LFSGGSTGLDD294 pKa = 2.85AYY296 pKa = 10.55DD297 pKa = 4.31AYY299 pKa = 10.88DD300 pKa = 3.4YY301 pKa = 11.25SLPVLLDD308 pKa = 3.19TGAAAWQVPSSRR320 pKa = 11.84YY321 pKa = 7.4TDD323 pKa = 3.95YY324 pKa = 10.84IAPAFPFVDD333 pKa = 3.3PLGYY337 pKa = 9.95CGCSHH342 pKa = 6.73RR343 pKa = 11.84QDD345 pKa = 5.6DD346 pKa = 3.9IQLTVTFGGEE356 pKa = 3.51IPIRR360 pKa = 11.84VPITDD365 pKa = 4.22FLVPIYY371 pKa = 9.62NTTTRR376 pKa = 11.84EE377 pKa = 4.2PIPYY381 pKa = 9.16TEE383 pKa = 5.93DD384 pKa = 3.38EE385 pKa = 4.55DD386 pKa = 4.27TCALLIVPAEE396 pKa = 3.81ATGYY400 pKa = 9.93GFQVLGDD407 pKa = 4.49AILRR411 pKa = 11.84SMYY414 pKa = 10.83VVFDD418 pKa = 4.61LDD420 pKa = 3.69NGQLSIAQAAEE431 pKa = 4.26DD432 pKa = 3.91NDD434 pKa = 4.12AGAGTSNPIPVPRR447 pKa = 11.84GPDD450 pKa = 3.74GIAAALSSASVGAAAEE466 pKa = 4.6DD467 pKa = 3.97YY468 pKa = 10.72RR469 pKa = 11.84SAEE472 pKa = 4.24STQTWSIAPLVTAASATGDD491 pKa = 3.87LSATTASRR499 pKa = 11.84TVGTATGSQAVPAGARR515 pKa = 11.84VSDD518 pKa = 4.14DD519 pKa = 3.67GVPGGDD525 pKa = 3.89DD526 pKa = 4.66GIGEE530 pKa = 4.96LIGVDD535 pKa = 5.59DD536 pKa = 6.72DD537 pKa = 6.85DD538 pKa = 7.88DD539 pKa = 7.42DD540 pKa = 7.55DD541 pKa = 7.05DD542 pKa = 7.16DD543 pKa = 7.12GDD545 pKa = 4.19GGSFGDD551 pKa = 4.51GGGEE555 pKa = 4.09ADD557 pKa = 4.45GSTSDD562 pKa = 4.06GSSSSSSSSGDD573 pKa = 3.29SSSTSSGAAAAVKK586 pKa = 10.4GGLSLGPDD594 pKa = 2.84WRR596 pKa = 11.84GWWVSGIVVLGVWVGMGVMLL616 pKa = 5.44
MM1 pKa = 7.36PRR3 pKa = 11.84SHH5 pKa = 6.56GRR7 pKa = 11.84RR8 pKa = 11.84TTAVLLTAAAAAATTVSALPEE29 pKa = 3.58IAITKK34 pKa = 10.04RR35 pKa = 11.84DD36 pKa = 3.7GEE38 pKa = 4.41LKK40 pKa = 10.14ASEE43 pKa = 4.14RR44 pKa = 11.84ARR46 pKa = 11.84GVEE49 pKa = 3.99KK50 pKa = 10.56RR51 pKa = 11.84AADD54 pKa = 3.55GNGTVEE60 pKa = 4.25TNVFDD65 pKa = 3.62VRR67 pKa = 11.84TWSTGGAYY75 pKa = 9.44YY76 pKa = 11.29ANVTVGTPPQPQVVILDD93 pKa = 3.84TGSSDD98 pKa = 4.8LYY100 pKa = 11.05FDD102 pKa = 4.79AADD105 pKa = 4.62APTCSLTGKK114 pKa = 8.35YY115 pKa = 9.41ACKK118 pKa = 10.43GGTFSPDD125 pKa = 2.75NSSTYY130 pKa = 10.34QVVDD134 pKa = 4.01PFPAFDD140 pKa = 3.8TLFGDD145 pKa = 4.74GSSATGEE152 pKa = 4.16YY153 pKa = 11.18GEE155 pKa = 4.52DD156 pKa = 3.16TVGIGDD162 pKa = 3.57VRR164 pKa = 11.84IEE166 pKa = 3.93NVQFGLANSLYY177 pKa = 8.45ITTGYY182 pKa = 11.18AVGLMGLGYY191 pKa = 10.34SYY193 pKa = 11.08IEE195 pKa = 5.15AIDD198 pKa = 4.02DD199 pKa = 4.39TEE201 pKa = 4.28DD202 pKa = 3.6LYY204 pKa = 10.93PNIPEE209 pKa = 4.05VLRR212 pKa = 11.84ATGVINSRR220 pKa = 11.84LYY222 pKa = 10.61SVYY225 pKa = 10.9LNDD228 pKa = 4.98LSDD231 pKa = 3.33ISGTILFGGIDD242 pKa = 3.61PSKK245 pKa = 9.79YY246 pKa = 9.3TGDD249 pKa = 3.53LTTLNILPDD258 pKa = 4.11VISGGISMFLTTVTDD273 pKa = 4.33LSITTDD279 pKa = 3.39GEE281 pKa = 4.35TADD284 pKa = 4.46LFSGGSTGLDD294 pKa = 2.85AYY296 pKa = 10.55DD297 pKa = 4.31AYY299 pKa = 10.88DD300 pKa = 3.4YY301 pKa = 11.25SLPVLLDD308 pKa = 3.19TGAAAWQVPSSRR320 pKa = 11.84YY321 pKa = 7.4TDD323 pKa = 3.95YY324 pKa = 10.84IAPAFPFVDD333 pKa = 3.3PLGYY337 pKa = 9.95CGCSHH342 pKa = 6.73RR343 pKa = 11.84QDD345 pKa = 5.6DD346 pKa = 3.9IQLTVTFGGEE356 pKa = 3.51IPIRR360 pKa = 11.84VPITDD365 pKa = 4.22FLVPIYY371 pKa = 9.62NTTTRR376 pKa = 11.84EE377 pKa = 4.2PIPYY381 pKa = 9.16TEE383 pKa = 5.93DD384 pKa = 3.38EE385 pKa = 4.55DD386 pKa = 4.27TCALLIVPAEE396 pKa = 3.81ATGYY400 pKa = 9.93GFQVLGDD407 pKa = 4.49AILRR411 pKa = 11.84SMYY414 pKa = 10.83VVFDD418 pKa = 4.61LDD420 pKa = 3.69NGQLSIAQAAEE431 pKa = 4.26DD432 pKa = 3.91NDD434 pKa = 4.12AGAGTSNPIPVPRR447 pKa = 11.84GPDD450 pKa = 3.74GIAAALSSASVGAAAEE466 pKa = 4.6DD467 pKa = 3.97YY468 pKa = 10.72RR469 pKa = 11.84SAEE472 pKa = 4.24STQTWSIAPLVTAASATGDD491 pKa = 3.87LSATTASRR499 pKa = 11.84TVGTATGSQAVPAGARR515 pKa = 11.84VSDD518 pKa = 4.14DD519 pKa = 3.67GVPGGDD525 pKa = 3.89DD526 pKa = 4.66GIGEE530 pKa = 4.96LIGVDD535 pKa = 5.59DD536 pKa = 6.72DD537 pKa = 6.85DD538 pKa = 7.88DD539 pKa = 7.42DD540 pKa = 7.55DD541 pKa = 7.05DD542 pKa = 7.16DD543 pKa = 7.12GDD545 pKa = 4.19GGSFGDD551 pKa = 4.51GGGEE555 pKa = 4.09ADD557 pKa = 4.45GSTSDD562 pKa = 4.06GSSSSSSSSGDD573 pKa = 3.29SSSTSSGAAAAVKK586 pKa = 10.4GGLSLGPDD594 pKa = 2.84WRR596 pKa = 11.84GWWVSGIVVLGVWVGMGVMLL616 pKa = 5.44
Molecular weight: 63.36 kDa
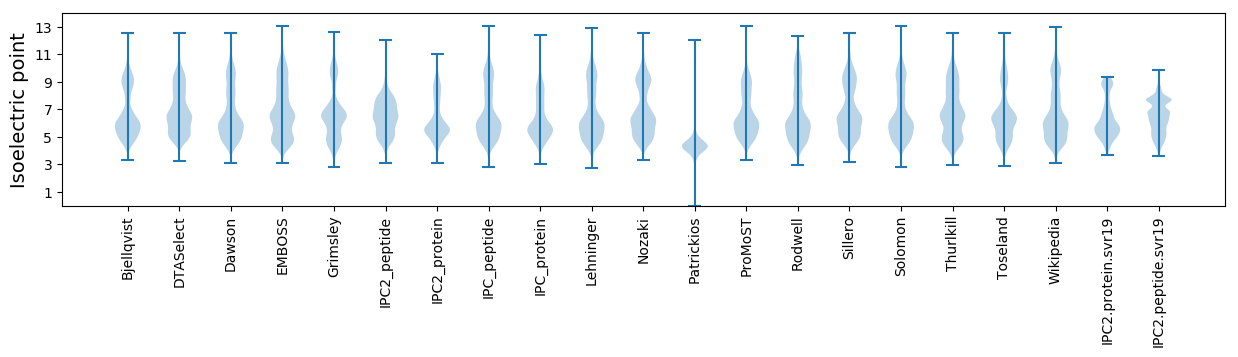
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Z5T3I2|A0A1Z5T3I2_HORWE MFS domain-containing protein OS=Hortaea werneckii EXF-2000 OX=1157616 GN=BTJ68_12399 PE=3 SV=1
MM1 pKa = 7.88PSHH4 pKa = 6.89KK5 pKa = 10.44SFRR8 pKa = 11.84TKK10 pKa = 10.65VKK12 pKa = 9.82LAKK15 pKa = 9.79AQKK18 pKa = 8.73QNRR21 pKa = 11.84PIPQWIRR28 pKa = 11.84LRR30 pKa = 11.84TGNTIRR36 pKa = 11.84YY37 pKa = 5.79NAKK40 pKa = 8.89RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 4.15WRR45 pKa = 11.84KK46 pKa = 7.38TRR48 pKa = 11.84IGII51 pKa = 4.0
MM1 pKa = 7.88PSHH4 pKa = 6.89KK5 pKa = 10.44SFRR8 pKa = 11.84TKK10 pKa = 10.65VKK12 pKa = 9.82LAKK15 pKa = 9.79AQKK18 pKa = 8.73QNRR21 pKa = 11.84PIPQWIRR28 pKa = 11.84LRR30 pKa = 11.84TGNTIRR36 pKa = 11.84YY37 pKa = 5.79NAKK40 pKa = 8.89RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 4.15WRR45 pKa = 11.84KK46 pKa = 7.38TRR48 pKa = 11.84IGII51 pKa = 4.0
Molecular weight: 6.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
8020772 |
50 |
4939 |
518.7 |
57.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.125 ± 0.015 | 1.125 ± 0.006 |
5.735 ± 0.015 | 6.461 ± 0.019 |
3.539 ± 0.013 | 7.48 ± 0.021 |
2.378 ± 0.008 | 4.318 ± 0.013 |
4.859 ± 0.018 | 8.43 ± 0.021 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.307 ± 0.008 | 3.646 ± 0.009 |
6.092 ± 0.021 | 4.475 ± 0.016 |
6.071 ± 0.015 | 8.055 ± 0.023 |
5.857 ± 0.016 | 5.874 ± 0.015 |
1.415 ± 0.007 | 2.756 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
