
Streptomyces glauciniger
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces
Average proteome isoelectric point is 6.46
Get precalculated fractions of proteins
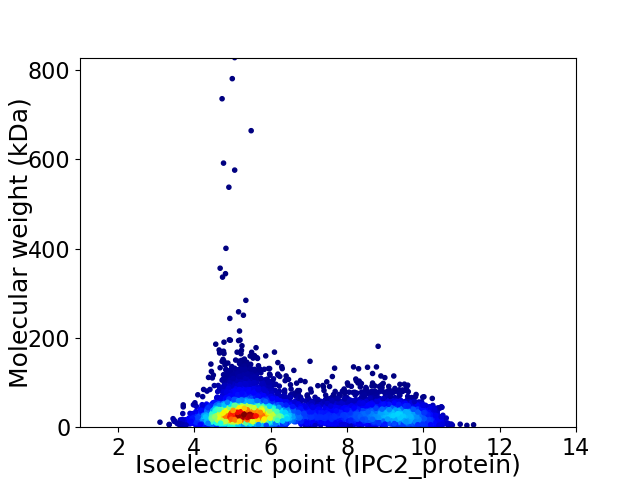
Virtual 2D-PAGE plot for 8673 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A239H820|A0A239H820_9ACTN Sensor histidine kinase regulating citrate/malate metabolism OS=Streptomyces glauciniger OX=235986 GN=SAMN05216252_108234 PE=4 SV=1
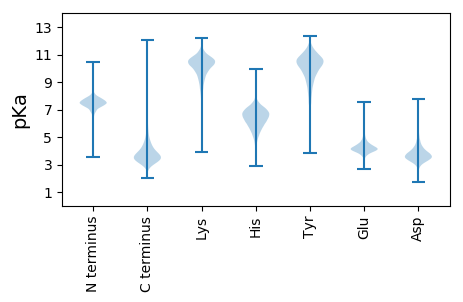
MM1 pKa = 7.22SHH3 pKa = 7.33DD4 pKa = 4.5PEE6 pKa = 5.74NPTPRR11 pKa = 11.84DD12 pKa = 3.36AASEE16 pKa = 4.19NPAAAAPSSSEE27 pKa = 3.86APADD31 pKa = 3.7ARR33 pKa = 11.84PTPDD37 pKa = 3.63FEE39 pKa = 5.51LPAPAPADD47 pKa = 3.71AVPADD52 pKa = 4.22AGSGADD58 pKa = 3.39SAGDD62 pKa = 3.68AGPGDD67 pKa = 4.07GAQADD72 pKa = 4.13VTAADD77 pKa = 4.38APAAPAGDD85 pKa = 4.63EE86 pKa = 4.13DD87 pKa = 4.42TKK89 pKa = 11.34VPGTVAPEE97 pKa = 3.96DD98 pKa = 3.36AAAPADD104 pKa = 4.01APGGAAALGDD114 pKa = 4.28APPVPTAHH122 pKa = 7.67PAPAPPPAGVPGAPDD137 pKa = 3.49PWAPPVPVAPGYY149 pKa = 8.28GQPGAHH155 pKa = 6.72AGYY158 pKa = 10.14NPWAPPIPQPPQEE171 pKa = 4.16QVTLTKK177 pKa = 10.51RR178 pKa = 11.84PVPEE182 pKa = 4.59DD183 pKa = 3.3AQAPGPAQQPRR194 pKa = 11.84DD195 pKa = 3.57PWAPPAPDD203 pKa = 3.42AAVTLPPQPPHH214 pKa = 5.88ATFPGYY220 pKa = 9.37PGPPGPPGYY229 pKa = 8.82PAYY232 pKa = 8.6PAHH235 pKa = 7.07PGYY238 pKa = 9.53PGYY241 pKa = 9.56PGYY244 pKa = 9.55PGQPGYY250 pKa = 9.77PGFGMGPYY258 pKa = 9.89GGWAPPVARR267 pKa = 11.84NGVGLTAMILGIVGAVLAISCFGAFLGLPLGIAAIACGIVGLRR310 pKa = 11.84IARR313 pKa = 11.84RR314 pKa = 11.84GEE316 pKa = 3.87ATNRR320 pKa = 11.84PQALTGLILGIISVVLAGGMIALVVGGIDD349 pKa = 3.49EE350 pKa = 4.56GWFDD354 pKa = 4.87GPGGDD359 pKa = 5.38DD360 pKa = 4.29LVTSRR365 pKa = 11.84NADD368 pKa = 2.94WEE370 pKa = 4.68TPLDD374 pKa = 3.93AEE376 pKa = 4.63GTALYY381 pKa = 10.57DD382 pKa = 4.16DD383 pKa = 5.01GVHH386 pKa = 5.19VTLSDD391 pKa = 3.73VQRR394 pKa = 11.84TTALPNSMLEE404 pKa = 4.05GGTAVTFSIEE414 pKa = 4.23LEE416 pKa = 4.16NTGDD420 pKa = 3.6DD421 pKa = 3.69TADD424 pKa = 3.45LSEE427 pKa = 4.89SEE429 pKa = 3.8ITAYY433 pKa = 10.55GDD435 pKa = 3.55DD436 pKa = 5.17FEE438 pKa = 7.37DD439 pKa = 3.39EE440 pKa = 4.35TLRR443 pKa = 11.84DD444 pKa = 3.46LTTGVGLPDD453 pKa = 3.35EE454 pKa = 4.63LAPGEE459 pKa = 4.24RR460 pKa = 11.84ATRR463 pKa = 11.84DD464 pKa = 2.96ITVVVPEE471 pKa = 4.41GDD473 pKa = 3.22EE474 pKa = 5.04DD475 pKa = 3.96GTFQVEE481 pKa = 4.31VAPGYY486 pKa = 10.96AYY488 pKa = 10.21DD489 pKa = 3.68YY490 pKa = 9.84TYY492 pKa = 11.2WNLDD496 pKa = 3.33VPP498 pKa = 4.46
MM1 pKa = 7.22SHH3 pKa = 7.33DD4 pKa = 4.5PEE6 pKa = 5.74NPTPRR11 pKa = 11.84DD12 pKa = 3.36AASEE16 pKa = 4.19NPAAAAPSSSEE27 pKa = 3.86APADD31 pKa = 3.7ARR33 pKa = 11.84PTPDD37 pKa = 3.63FEE39 pKa = 5.51LPAPAPADD47 pKa = 3.71AVPADD52 pKa = 4.22AGSGADD58 pKa = 3.39SAGDD62 pKa = 3.68AGPGDD67 pKa = 4.07GAQADD72 pKa = 4.13VTAADD77 pKa = 4.38APAAPAGDD85 pKa = 4.63EE86 pKa = 4.13DD87 pKa = 4.42TKK89 pKa = 11.34VPGTVAPEE97 pKa = 3.96DD98 pKa = 3.36AAAPADD104 pKa = 4.01APGGAAALGDD114 pKa = 4.28APPVPTAHH122 pKa = 7.67PAPAPPPAGVPGAPDD137 pKa = 3.49PWAPPVPVAPGYY149 pKa = 8.28GQPGAHH155 pKa = 6.72AGYY158 pKa = 10.14NPWAPPIPQPPQEE171 pKa = 4.16QVTLTKK177 pKa = 10.51RR178 pKa = 11.84PVPEE182 pKa = 4.59DD183 pKa = 3.3AQAPGPAQQPRR194 pKa = 11.84DD195 pKa = 3.57PWAPPAPDD203 pKa = 3.42AAVTLPPQPPHH214 pKa = 5.88ATFPGYY220 pKa = 9.37PGPPGPPGYY229 pKa = 8.82PAYY232 pKa = 8.6PAHH235 pKa = 7.07PGYY238 pKa = 9.53PGYY241 pKa = 9.56PGYY244 pKa = 9.55PGQPGYY250 pKa = 9.77PGFGMGPYY258 pKa = 9.89GGWAPPVARR267 pKa = 11.84NGVGLTAMILGIVGAVLAISCFGAFLGLPLGIAAIACGIVGLRR310 pKa = 11.84IARR313 pKa = 11.84RR314 pKa = 11.84GEE316 pKa = 3.87ATNRR320 pKa = 11.84PQALTGLILGIISVVLAGGMIALVVGGIDD349 pKa = 3.49EE350 pKa = 4.56GWFDD354 pKa = 4.87GPGGDD359 pKa = 5.38DD360 pKa = 4.29LVTSRR365 pKa = 11.84NADD368 pKa = 2.94WEE370 pKa = 4.68TPLDD374 pKa = 3.93AEE376 pKa = 4.63GTALYY381 pKa = 10.57DD382 pKa = 4.16DD383 pKa = 5.01GVHH386 pKa = 5.19VTLSDD391 pKa = 3.73VQRR394 pKa = 11.84TTALPNSMLEE404 pKa = 4.05GGTAVTFSIEE414 pKa = 4.23LEE416 pKa = 4.16NTGDD420 pKa = 3.6DD421 pKa = 3.69TADD424 pKa = 3.45LSEE427 pKa = 4.89SEE429 pKa = 3.8ITAYY433 pKa = 10.55GDD435 pKa = 3.55DD436 pKa = 5.17FEE438 pKa = 7.37DD439 pKa = 3.39EE440 pKa = 4.35TLRR443 pKa = 11.84DD444 pKa = 3.46LTTGVGLPDD453 pKa = 3.35EE454 pKa = 4.63LAPGEE459 pKa = 4.24RR460 pKa = 11.84ATRR463 pKa = 11.84DD464 pKa = 2.96ITVVVPEE471 pKa = 4.41GDD473 pKa = 3.22EE474 pKa = 5.04DD475 pKa = 3.96GTFQVEE481 pKa = 4.31VAPGYY486 pKa = 10.96AYY488 pKa = 10.21DD489 pKa = 3.68YY490 pKa = 9.84TYY492 pKa = 11.2WNLDD496 pKa = 3.33VPP498 pKa = 4.46
Molecular weight: 49.99 kDa
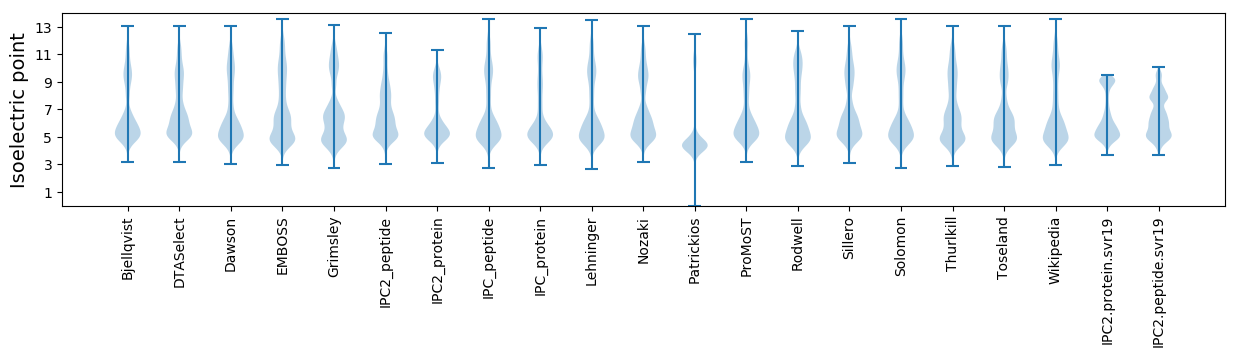
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A239IEG9|A0A239IEG9_9ACTN Predicted oxidoreductase OS=Streptomyces glauciniger OX=235986 GN=SAMN05216252_110124 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILATRR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.88GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILATRR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.88GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
Molecular weight: 5.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2848599 |
24 |
7714 |
328.4 |
35.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.003 ± 0.041 | 0.776 ± 0.008 |
5.982 ± 0.022 | 5.553 ± 0.027 |
2.703 ± 0.015 | 9.733 ± 0.031 |
2.334 ± 0.013 | 2.958 ± 0.016 |
1.887 ± 0.021 | 10.322 ± 0.034 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.703 ± 0.011 | 1.681 ± 0.014 |
6.203 ± 0.027 | 2.62 ± 0.015 |
8.273 ± 0.029 | 4.966 ± 0.021 |
6.053 ± 0.027 | 8.655 ± 0.028 |
1.546 ± 0.012 | 2.05 ± 0.014 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
