
Saccharothrix espanaensis (strain ATCC 51144 / DSM 44229 / JCM 9112 / NBRC 15066 / NRRL 15764)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Pseudonocardiales; Pseudonocardiaceae; Saccharothrix; Saccharothrix espanaensis
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
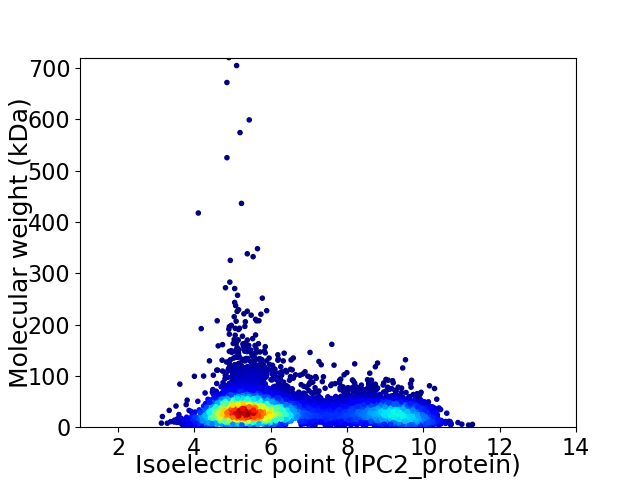
Virtual 2D-PAGE plot for 8423 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K0K5K3|K0K5K3_SACES Guanylate kinase OS=Saccharothrix espanaensis (strain ATCC 51144 / DSM 44229 / JCM 9112 / NBRC 15066 / NRRL 15764) OX=1179773 GN=gmk PE=3 SV=1
MM1 pKa = 7.6SVEE4 pKa = 3.93TFQRR8 pKa = 11.84SRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84GGWHH17 pKa = 6.56RR18 pKa = 11.84RR19 pKa = 11.84LAMTAAVSVASAGLLAVGAQPASAAVVTHH48 pKa = 6.3PFAYY52 pKa = 9.89VANSGANTVTAYY64 pKa = 10.23DD65 pKa = 3.46ATTATVASTIVVGSTPLGLVATPDD89 pKa = 3.39GARR92 pKa = 11.84VYY94 pKa = 9.9VAKK97 pKa = 10.77LSGGVSVIDD106 pKa = 3.61TAAGAVVASIAVGSQPIGIAITVDD130 pKa = 3.09GTRR133 pKa = 11.84ILVVNSGSNSVSMIDD148 pKa = 3.33TATNAVTATVTVGANPTSVAISPDD172 pKa = 3.03GATAYY177 pKa = 7.83VTNDD181 pKa = 2.63ASGTVSVIDD190 pKa = 3.61TATATVTGTINLGLTQPAGVAITPDD215 pKa = 3.21GATVYY220 pKa = 9.7VARR223 pKa = 11.84YY224 pKa = 8.3GAGTVAVIDD233 pKa = 3.87TATNAVTGSITVGAVPSGVAVSPDD257 pKa = 3.0GTRR260 pKa = 11.84AYY262 pKa = 9.76VANIGPDD269 pKa = 3.76TISVIDD275 pKa = 3.8TATDD279 pKa = 3.38TVTATVPVGSNPSRR293 pKa = 11.84VAVTDD298 pKa = 4.1DD299 pKa = 3.23GSTVYY304 pKa = 9.39ATNNGAGSVAAINTATNTVTTTFAVGSGPFGVTTANVVQPRR345 pKa = 11.84VTAISPTTGPTAGGTSVTITGTSLAAATAVRR376 pKa = 11.84FGATPATSFTAVNDD390 pKa = 3.91TTVTATAPAHH400 pKa = 5.9AAGPVDD406 pKa = 3.78VTVTTPTGVSTTSAADD422 pKa = 3.08QYY424 pKa = 11.03TYY426 pKa = 11.24LDD428 pKa = 4.1PAPVISAVDD437 pKa = 3.73PATGPAAGGTPVTITGTHH455 pKa = 4.92FTGATAVAFGGTPATSFTVVNDD477 pKa = 3.57TSITATAPAGTAGTVDD493 pKa = 3.44VTVTNAGGTSAPGSGYY509 pKa = 9.42TYY511 pKa = 10.27VAAPAVTALDD521 pKa = 3.91PTSGPTAGGTAVAITGTALTGTTAVRR547 pKa = 11.84FDD549 pKa = 3.98GVDD552 pKa = 3.03ATSFTVTSSTSITAVAPAHH571 pKa = 5.96SAGTVDD577 pKa = 3.43VTVTTVGGTSAATASSRR594 pKa = 11.84YY595 pKa = 7.93TYY597 pKa = 11.4ADD599 pKa = 3.33ADD601 pKa = 4.04LSVSVTDD608 pKa = 3.64SADD611 pKa = 3.59PVSLGGDD618 pKa = 3.72PYY620 pKa = 10.8TYY622 pKa = 10.66TVTAANAGPSTATGVTVSTTLSGASTTINSATPSQGSCTTGAATVSCTLGTIAGSGSASVTISVTPSTSGTATATAAVSATEE704 pKa = 4.21PDD706 pKa = 3.82PVPANNTDD714 pKa = 3.8TEE716 pKa = 4.57STTVDD721 pKa = 3.1NALGCTIVGTAGNDD735 pKa = 3.55SLNGTNGNDD744 pKa = 3.81VICGLGGNDD753 pKa = 4.5TITGGNGTDD762 pKa = 3.85TIHH765 pKa = 7.2AGTGDD770 pKa = 3.74DD771 pKa = 3.99VLVGGNGADD780 pKa = 3.33TLYY783 pKa = 11.23GGLGNDD789 pKa = 3.46TSKK792 pKa = 11.54GEE794 pKa = 4.11SLLDD798 pKa = 3.53SLLGLFDD805 pKa = 5.09NGDD808 pKa = 3.62DD809 pKa = 4.53TIDD812 pKa = 4.03GGPGNDD818 pKa = 4.46NLDD821 pKa = 3.81GQNGNDD827 pKa = 3.72TLIDD831 pKa = 3.68HH832 pKa = 7.56DD833 pKa = 4.74GTDD836 pKa = 3.52TMAGSLGNDD845 pKa = 3.78TIDD848 pKa = 3.45VQDD851 pKa = 4.96GIGGDD856 pKa = 3.77TANGGLGTDD865 pKa = 3.33TCTVDD870 pKa = 4.72GGDD873 pKa = 3.74TTSSCC878 pKa = 4.44
MM1 pKa = 7.6SVEE4 pKa = 3.93TFQRR8 pKa = 11.84SRR10 pKa = 11.84RR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84GGWHH17 pKa = 6.56RR18 pKa = 11.84RR19 pKa = 11.84LAMTAAVSVASAGLLAVGAQPASAAVVTHH48 pKa = 6.3PFAYY52 pKa = 9.89VANSGANTVTAYY64 pKa = 10.23DD65 pKa = 3.46ATTATVASTIVVGSTPLGLVATPDD89 pKa = 3.39GARR92 pKa = 11.84VYY94 pKa = 9.9VAKK97 pKa = 10.77LSGGVSVIDD106 pKa = 3.61TAAGAVVASIAVGSQPIGIAITVDD130 pKa = 3.09GTRR133 pKa = 11.84ILVVNSGSNSVSMIDD148 pKa = 3.33TATNAVTATVTVGANPTSVAISPDD172 pKa = 3.03GATAYY177 pKa = 7.83VTNDD181 pKa = 2.63ASGTVSVIDD190 pKa = 3.61TATATVTGTINLGLTQPAGVAITPDD215 pKa = 3.21GATVYY220 pKa = 9.7VARR223 pKa = 11.84YY224 pKa = 8.3GAGTVAVIDD233 pKa = 3.87TATNAVTGSITVGAVPSGVAVSPDD257 pKa = 3.0GTRR260 pKa = 11.84AYY262 pKa = 9.76VANIGPDD269 pKa = 3.76TISVIDD275 pKa = 3.8TATDD279 pKa = 3.38TVTATVPVGSNPSRR293 pKa = 11.84VAVTDD298 pKa = 4.1DD299 pKa = 3.23GSTVYY304 pKa = 9.39ATNNGAGSVAAINTATNTVTTTFAVGSGPFGVTTANVVQPRR345 pKa = 11.84VTAISPTTGPTAGGTSVTITGTSLAAATAVRR376 pKa = 11.84FGATPATSFTAVNDD390 pKa = 3.91TTVTATAPAHH400 pKa = 5.9AAGPVDD406 pKa = 3.78VTVTTPTGVSTTSAADD422 pKa = 3.08QYY424 pKa = 11.03TYY426 pKa = 11.24LDD428 pKa = 4.1PAPVISAVDD437 pKa = 3.73PATGPAAGGTPVTITGTHH455 pKa = 4.92FTGATAVAFGGTPATSFTVVNDD477 pKa = 3.57TSITATAPAGTAGTVDD493 pKa = 3.44VTVTNAGGTSAPGSGYY509 pKa = 9.42TYY511 pKa = 10.27VAAPAVTALDD521 pKa = 3.91PTSGPTAGGTAVAITGTALTGTTAVRR547 pKa = 11.84FDD549 pKa = 3.98GVDD552 pKa = 3.03ATSFTVTSSTSITAVAPAHH571 pKa = 5.96SAGTVDD577 pKa = 3.43VTVTTVGGTSAATASSRR594 pKa = 11.84YY595 pKa = 7.93TYY597 pKa = 11.4ADD599 pKa = 3.33ADD601 pKa = 4.04LSVSVTDD608 pKa = 3.64SADD611 pKa = 3.59PVSLGGDD618 pKa = 3.72PYY620 pKa = 10.8TYY622 pKa = 10.66TVTAANAGPSTATGVTVSTTLSGASTTINSATPSQGSCTTGAATVSCTLGTIAGSGSASVTISVTPSTSGTATATAAVSATEE704 pKa = 4.21PDD706 pKa = 3.82PVPANNTDD714 pKa = 3.8TEE716 pKa = 4.57STTVDD721 pKa = 3.1NALGCTIVGTAGNDD735 pKa = 3.55SLNGTNGNDD744 pKa = 3.81VICGLGGNDD753 pKa = 4.5TITGGNGTDD762 pKa = 3.85TIHH765 pKa = 7.2AGTGDD770 pKa = 3.74DD771 pKa = 3.99VLVGGNGADD780 pKa = 3.33TLYY783 pKa = 11.23GGLGNDD789 pKa = 3.46TSKK792 pKa = 11.54GEE794 pKa = 4.11SLLDD798 pKa = 3.53SLLGLFDD805 pKa = 5.09NGDD808 pKa = 3.62DD809 pKa = 4.53TIDD812 pKa = 4.03GGPGNDD818 pKa = 4.46NLDD821 pKa = 3.81GQNGNDD827 pKa = 3.72TLIDD831 pKa = 3.68HH832 pKa = 7.56DD833 pKa = 4.74GTDD836 pKa = 3.52TMAGSLGNDD845 pKa = 3.78TIDD848 pKa = 3.45VQDD851 pKa = 4.96GIGGDD856 pKa = 3.77TANGGLGTDD865 pKa = 3.33TCTVDD870 pKa = 4.72GGDD873 pKa = 3.74TTSSCC878 pKa = 4.44
Molecular weight: 83.94 kDa
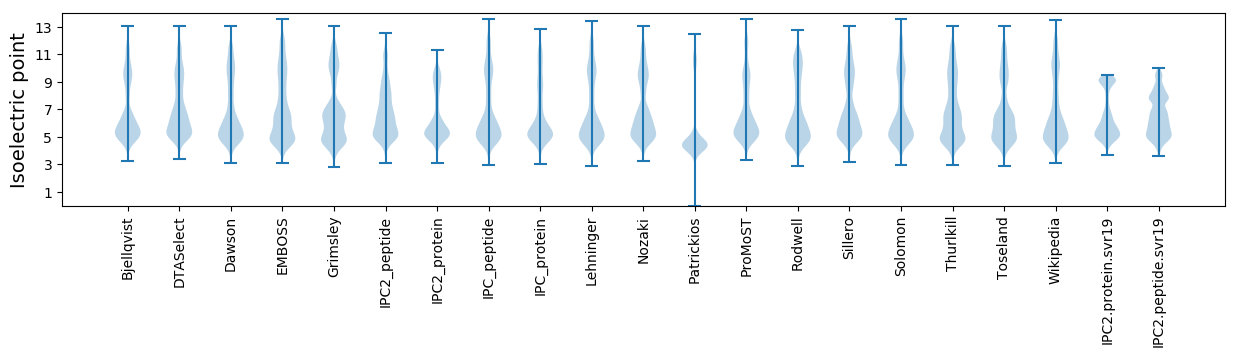
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K0KCC7|K0KCC7_SACES ABC-type transporter substrate-binding lipoprotein OS=Saccharothrix espanaensis (strain ATCC 51144 / DSM 44229 / JCM 9112 / NBRC 15066 / NRRL 15764) OX=1179773 GN=BN6_79590 PE=4 SV=1
MM1 pKa = 7.53SKK3 pKa = 10.53GKK5 pKa = 8.66RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84AKK17 pKa = 8.7THH19 pKa = 5.15GFRR22 pKa = 11.84LRR24 pKa = 11.84MRR26 pKa = 11.84TRR28 pKa = 11.84AGRR31 pKa = 11.84AILAARR37 pKa = 11.84RR38 pKa = 11.84SKK40 pKa = 10.77GRR42 pKa = 11.84QQLSAA47 pKa = 4.05
MM1 pKa = 7.53SKK3 pKa = 10.53GKK5 pKa = 8.66RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84AKK17 pKa = 8.7THH19 pKa = 5.15GFRR22 pKa = 11.84LRR24 pKa = 11.84MRR26 pKa = 11.84TRR28 pKa = 11.84AGRR31 pKa = 11.84AILAARR37 pKa = 11.84RR38 pKa = 11.84SKK40 pKa = 10.77GRR42 pKa = 11.84QQLSAA47 pKa = 4.05
Molecular weight: 5.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2787681 |
27 |
6716 |
331.0 |
35.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.441 ± 0.043 | 0.793 ± 0.008 |
6.286 ± 0.025 | 5.316 ± 0.021 |
2.796 ± 0.015 | 9.338 ± 0.027 |
2.349 ± 0.015 | 2.791 ± 0.018 |
1.853 ± 0.02 | 10.491 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.471 ± 0.011 | 1.773 ± 0.019 |
6.154 ± 0.026 | 2.573 ± 0.016 |
8.467 ± 0.031 | 4.895 ± 0.02 |
6.082 ± 0.037 | 9.716 ± 0.027 |
1.547 ± 0.011 | 1.869 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
