
Caenorhabditis remanei (Caenorhabditis vulgaris)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Nematoda; Chromadorea; Rhabditida; Rhabditina; Rhabditomorpha; Rhabditoidea; Rhabditidae; Peloderinae; Caenorhabditis
Average proteome isoelectric point is 6.78
Get precalculated fractions of proteins
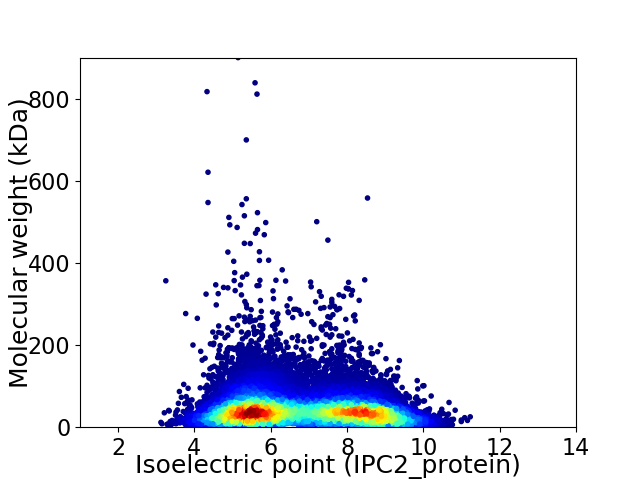
Virtual 2D-PAGE plot for 31252 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E3NCH3|E3NCH3_CAERE Uncharacterized protein OS=Caenorhabditis remanei OX=31234 GN=CRE_18208 PE=4 SV=1
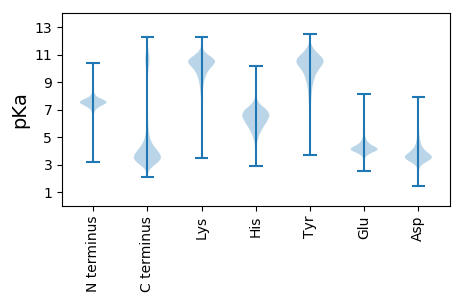
MM1 pKa = 7.57SDD3 pKa = 3.09EE4 pKa = 4.15EE5 pKa = 4.4EE6 pKa = 4.96YY7 pKa = 11.3YY8 pKa = 11.02DD9 pKa = 5.0EE10 pKa = 6.45DD11 pKa = 5.59GGGQDD16 pKa = 4.52DD17 pKa = 5.07QGGDD21 pKa = 3.65DD22 pKa = 4.03YY23 pKa = 11.52GYY25 pKa = 11.04SGDD28 pKa = 3.96DD29 pKa = 3.53FGGGGYY35 pKa = 10.32EE36 pKa = 5.81DD37 pKa = 4.42GNQYY41 pKa = 10.77DD42 pKa = 3.59QNQGYY47 pKa = 8.91DD48 pKa = 3.07QNNQYY53 pKa = 9.78GQGYY57 pKa = 7.95GQDD60 pKa = 3.83YY61 pKa = 10.65GNQGYY66 pKa = 10.49GNQQDD71 pKa = 4.29YY72 pKa = 11.6GNQQYY77 pKa = 9.89GQGGGGYY84 pKa = 9.41DD85 pKa = 4.41QEE87 pKa = 4.46YY88 pKa = 9.22QQNDD92 pKa = 3.85YY93 pKa = 11.38QGGGYY98 pKa = 9.88DD99 pKa = 3.53DD100 pKa = 3.84QEE102 pKa = 4.53QYY104 pKa = 11.29EE105 pKa = 4.5EE106 pKa = 4.84DD107 pKa = 3.66PEE109 pKa = 4.25EE110 pKa = 4.64PEE112 pKa = 4.3EE113 pKa = 4.39EE114 pKa = 4.14EE115 pKa = 4.79DD116 pKa = 5.56PEE118 pKa = 5.79PEE120 pKa = 4.01DD121 pKa = 4.0QEE123 pKa = 4.27VDD125 pKa = 3.33EE126 pKa = 4.95SEE128 pKa = 4.2EE129 pKa = 4.21DD130 pKa = 3.75VVEE133 pKa = 4.05TSEE136 pKa = 4.52DD137 pKa = 3.53VEE139 pKa = 4.35EE140 pKa = 4.82EE141 pKa = 4.16EE142 pKa = 5.48EE143 pKa = 4.15EE144 pKa = 4.36DD145 pKa = 4.75DD146 pKa = 6.3GYY148 pKa = 9.24TQVAAHH154 pKa = 6.62TNQASDD160 pKa = 3.67GNGGDD165 pKa = 4.64GNNADD170 pKa = 4.54HH171 pKa = 7.19GLGSLFKK178 pKa = 10.85GAMDD182 pKa = 4.28GFGGGGFDD190 pKa = 4.38GVVGSIGSLISQGGGDD206 pKa = 3.7STGGLTNVFSSGGMEE221 pKa = 4.23SIVGNLISSASHH233 pKa = 5.09QFFGINPSTGAIIGAIAGNIIFQMGGQNNSLSSIGKK269 pKa = 9.12VVLDD273 pKa = 4.54NIISGKK279 pKa = 8.45FKK281 pKa = 10.81RR282 pKa = 11.84DD283 pKa = 3.2IQPFTPGGTVPGLGFQQQFQTINFQQEE310 pKa = 3.75RR311 pKa = 11.84QRR313 pKa = 11.84CLEE316 pKa = 3.71QRR318 pKa = 11.84ILFEE322 pKa = 5.48DD323 pKa = 3.86PQFPANDD330 pKa = 3.04SSLYY334 pKa = 10.21YY335 pKa = 9.17KK336 pKa = 9.69TRR338 pKa = 11.84PDD340 pKa = 3.65EE341 pKa = 4.7PIVWKK346 pKa = 10.5RR347 pKa = 11.84PGEE350 pKa = 4.06IYY352 pKa = 10.41EE353 pKa = 4.1NPQLIVGEE361 pKa = 4.23KK362 pKa = 10.33SRR364 pKa = 11.84FDD366 pKa = 3.83VKK368 pKa = 10.73QGALGDD374 pKa = 4.03CWLLAAVANLTLRR387 pKa = 11.84DD388 pKa = 3.59EE389 pKa = 4.3LFYY392 pKa = 11.02RR393 pKa = 11.84VVPPDD398 pKa = 3.15QSFTEE403 pKa = 4.28NYY405 pKa = 10.35AGIFHH410 pKa = 6.35FQFWRR415 pKa = 11.84YY416 pKa = 7.49GQWVDD421 pKa = 3.97VVIDD425 pKa = 3.94DD426 pKa = 5.2RR427 pKa = 11.84LPTVDD432 pKa = 5.42GRR434 pKa = 11.84LCYY437 pKa = 9.71MRR439 pKa = 11.84SQEE442 pKa = 4.75NNEE445 pKa = 3.83FWSALLEE452 pKa = 3.89KK453 pKa = 10.25AYY455 pKa = 10.72AKK457 pKa = 10.62LYY459 pKa = 10.57GGYY462 pKa = 9.4EE463 pKa = 4.08HH464 pKa = 7.66LDD466 pKa = 3.47GGTTAEE472 pKa = 4.25ALEE475 pKa = 4.92DD476 pKa = 3.74FTGGLTEE483 pKa = 5.66FFDD486 pKa = 4.88LEE488 pKa = 4.58KK489 pKa = 10.84GDD491 pKa = 3.99KK492 pKa = 9.35STILAMLVRR501 pKa = 11.84GMQMGSLFGCSIDD514 pKa = 3.61ADD516 pKa = 4.18EE517 pKa = 4.67NVKK520 pKa = 9.91EE521 pKa = 4.07AQLTNGLVRR530 pKa = 11.84GHH532 pKa = 7.28AYY534 pKa = 10.55SITAIQTVNTYY545 pKa = 10.57SPSPLNQWNPTPPISLQFSS564 pKa = 3.69
MM1 pKa = 7.57SDD3 pKa = 3.09EE4 pKa = 4.15EE5 pKa = 4.4EE6 pKa = 4.96YY7 pKa = 11.3YY8 pKa = 11.02DD9 pKa = 5.0EE10 pKa = 6.45DD11 pKa = 5.59GGGQDD16 pKa = 4.52DD17 pKa = 5.07QGGDD21 pKa = 3.65DD22 pKa = 4.03YY23 pKa = 11.52GYY25 pKa = 11.04SGDD28 pKa = 3.96DD29 pKa = 3.53FGGGGYY35 pKa = 10.32EE36 pKa = 5.81DD37 pKa = 4.42GNQYY41 pKa = 10.77DD42 pKa = 3.59QNQGYY47 pKa = 8.91DD48 pKa = 3.07QNNQYY53 pKa = 9.78GQGYY57 pKa = 7.95GQDD60 pKa = 3.83YY61 pKa = 10.65GNQGYY66 pKa = 10.49GNQQDD71 pKa = 4.29YY72 pKa = 11.6GNQQYY77 pKa = 9.89GQGGGGYY84 pKa = 9.41DD85 pKa = 4.41QEE87 pKa = 4.46YY88 pKa = 9.22QQNDD92 pKa = 3.85YY93 pKa = 11.38QGGGYY98 pKa = 9.88DD99 pKa = 3.53DD100 pKa = 3.84QEE102 pKa = 4.53QYY104 pKa = 11.29EE105 pKa = 4.5EE106 pKa = 4.84DD107 pKa = 3.66PEE109 pKa = 4.25EE110 pKa = 4.64PEE112 pKa = 4.3EE113 pKa = 4.39EE114 pKa = 4.14EE115 pKa = 4.79DD116 pKa = 5.56PEE118 pKa = 5.79PEE120 pKa = 4.01DD121 pKa = 4.0QEE123 pKa = 4.27VDD125 pKa = 3.33EE126 pKa = 4.95SEE128 pKa = 4.2EE129 pKa = 4.21DD130 pKa = 3.75VVEE133 pKa = 4.05TSEE136 pKa = 4.52DD137 pKa = 3.53VEE139 pKa = 4.35EE140 pKa = 4.82EE141 pKa = 4.16EE142 pKa = 5.48EE143 pKa = 4.15EE144 pKa = 4.36DD145 pKa = 4.75DD146 pKa = 6.3GYY148 pKa = 9.24TQVAAHH154 pKa = 6.62TNQASDD160 pKa = 3.67GNGGDD165 pKa = 4.64GNNADD170 pKa = 4.54HH171 pKa = 7.19GLGSLFKK178 pKa = 10.85GAMDD182 pKa = 4.28GFGGGGFDD190 pKa = 4.38GVVGSIGSLISQGGGDD206 pKa = 3.7STGGLTNVFSSGGMEE221 pKa = 4.23SIVGNLISSASHH233 pKa = 5.09QFFGINPSTGAIIGAIAGNIIFQMGGQNNSLSSIGKK269 pKa = 9.12VVLDD273 pKa = 4.54NIISGKK279 pKa = 8.45FKK281 pKa = 10.81RR282 pKa = 11.84DD283 pKa = 3.2IQPFTPGGTVPGLGFQQQFQTINFQQEE310 pKa = 3.75RR311 pKa = 11.84QRR313 pKa = 11.84CLEE316 pKa = 3.71QRR318 pKa = 11.84ILFEE322 pKa = 5.48DD323 pKa = 3.86PQFPANDD330 pKa = 3.04SSLYY334 pKa = 10.21YY335 pKa = 9.17KK336 pKa = 9.69TRR338 pKa = 11.84PDD340 pKa = 3.65EE341 pKa = 4.7PIVWKK346 pKa = 10.5RR347 pKa = 11.84PGEE350 pKa = 4.06IYY352 pKa = 10.41EE353 pKa = 4.1NPQLIVGEE361 pKa = 4.23KK362 pKa = 10.33SRR364 pKa = 11.84FDD366 pKa = 3.83VKK368 pKa = 10.73QGALGDD374 pKa = 4.03CWLLAAVANLTLRR387 pKa = 11.84DD388 pKa = 3.59EE389 pKa = 4.3LFYY392 pKa = 11.02RR393 pKa = 11.84VVPPDD398 pKa = 3.15QSFTEE403 pKa = 4.28NYY405 pKa = 10.35AGIFHH410 pKa = 6.35FQFWRR415 pKa = 11.84YY416 pKa = 7.49GQWVDD421 pKa = 3.97VVIDD425 pKa = 3.94DD426 pKa = 5.2RR427 pKa = 11.84LPTVDD432 pKa = 5.42GRR434 pKa = 11.84LCYY437 pKa = 9.71MRR439 pKa = 11.84SQEE442 pKa = 4.75NNEE445 pKa = 3.83FWSALLEE452 pKa = 3.89KK453 pKa = 10.25AYY455 pKa = 10.72AKK457 pKa = 10.62LYY459 pKa = 10.57GGYY462 pKa = 9.4EE463 pKa = 4.08HH464 pKa = 7.66LDD466 pKa = 3.47GGTTAEE472 pKa = 4.25ALEE475 pKa = 4.92DD476 pKa = 3.74FTGGLTEE483 pKa = 5.66FFDD486 pKa = 4.88LEE488 pKa = 4.58KK489 pKa = 10.84GDD491 pKa = 3.99KK492 pKa = 9.35STILAMLVRR501 pKa = 11.84GMQMGSLFGCSIDD514 pKa = 3.61ADD516 pKa = 4.18EE517 pKa = 4.67NVKK520 pKa = 9.91EE521 pKa = 4.07AQLTNGLVRR530 pKa = 11.84GHH532 pKa = 7.28AYY534 pKa = 10.55SITAIQTVNTYY545 pKa = 10.57SPSPLNQWNPTPPISLQFSS564 pKa = 3.69
Molecular weight: 61.86 kDa
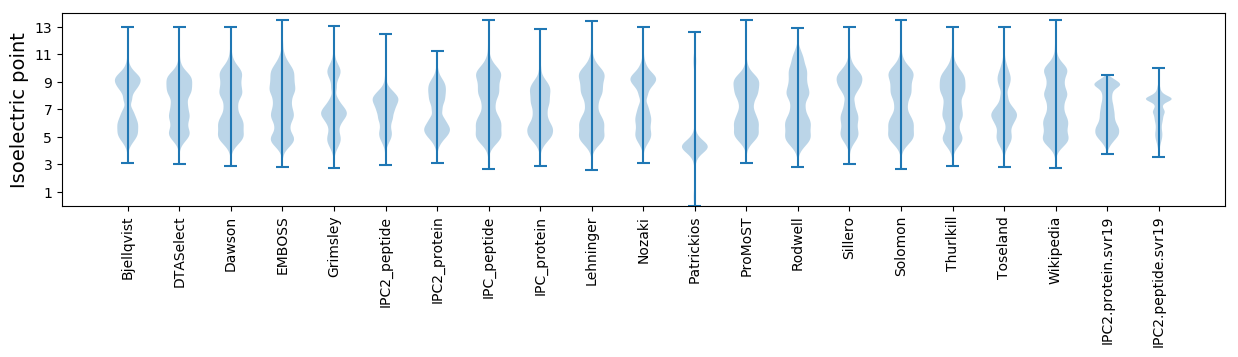
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E3LMT0|E3LMT0_CAERE Uncharacterized protein OS=Caenorhabditis remanei OX=31234 GN=CRE_28500 PE=4 SV=1
MM1 pKa = 7.28NGGRR5 pKa = 11.84KK6 pKa = 8.85RR7 pKa = 11.84RR8 pKa = 11.84FPVTVAACSILVLVISVVTAQQVFDD33 pKa = 4.65GNRR36 pKa = 11.84QPAFVRR42 pKa = 11.84RR43 pKa = 11.84QPAFQQQQPNQIQFQQNRR61 pKa = 11.84NFVLPSASPLRR72 pKa = 11.84HH73 pKa = 6.05RR74 pKa = 11.84NLFTRR79 pKa = 11.84QISHH83 pKa = 5.84RR84 pKa = 11.84HH85 pKa = 4.81RR86 pKa = 11.84PRR88 pKa = 11.84HH89 pKa = 5.42RR90 pKa = 11.84SSLRR94 pKa = 11.84KK95 pKa = 7.03TTHH98 pKa = 5.77QPQQAHH104 pKa = 4.73LTSRR108 pKa = 11.84RR109 pKa = 11.84VIPTLSHH116 pKa = 5.82KK117 pKa = 9.08RR118 pKa = 11.84TRR120 pKa = 11.84SRR122 pKa = 11.84TVVSSLSSRR131 pKa = 11.84TKK133 pKa = 9.63PNVTRR138 pKa = 11.84VVNANQRR145 pKa = 11.84TTTIKK150 pKa = 10.59SNNSRR155 pKa = 11.84RR156 pKa = 11.84QQQQFQPRR164 pKa = 11.84QPFMTQRR171 pKa = 11.84QQQFRR176 pKa = 11.84FQPQPQRR183 pKa = 11.84QQIFQRR189 pKa = 11.84VQRR192 pKa = 11.84VQNFQRR198 pKa = 11.84APIQNQQFFVRR209 pKa = 11.84QALNN213 pKa = 3.06
MM1 pKa = 7.28NGGRR5 pKa = 11.84KK6 pKa = 8.85RR7 pKa = 11.84RR8 pKa = 11.84FPVTVAACSILVLVISVVTAQQVFDD33 pKa = 4.65GNRR36 pKa = 11.84QPAFVRR42 pKa = 11.84RR43 pKa = 11.84QPAFQQQQPNQIQFQQNRR61 pKa = 11.84NFVLPSASPLRR72 pKa = 11.84HH73 pKa = 6.05RR74 pKa = 11.84NLFTRR79 pKa = 11.84QISHH83 pKa = 5.84RR84 pKa = 11.84HH85 pKa = 4.81RR86 pKa = 11.84PRR88 pKa = 11.84HH89 pKa = 5.42RR90 pKa = 11.84SSLRR94 pKa = 11.84KK95 pKa = 7.03TTHH98 pKa = 5.77QPQQAHH104 pKa = 4.73LTSRR108 pKa = 11.84RR109 pKa = 11.84VIPTLSHH116 pKa = 5.82KK117 pKa = 9.08RR118 pKa = 11.84TRR120 pKa = 11.84SRR122 pKa = 11.84TVVSSLSSRR131 pKa = 11.84TKK133 pKa = 9.63PNVTRR138 pKa = 11.84VVNANQRR145 pKa = 11.84TTTIKK150 pKa = 10.59SNNSRR155 pKa = 11.84RR156 pKa = 11.84QQQQFQPRR164 pKa = 11.84QPFMTQRR171 pKa = 11.84QQQFRR176 pKa = 11.84FQPQPQRR183 pKa = 11.84QQIFQRR189 pKa = 11.84VQRR192 pKa = 11.84VQNFQRR198 pKa = 11.84APIQNQQFFVRR209 pKa = 11.84QALNN213 pKa = 3.06
Molecular weight: 25.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
12547433 |
9 |
13921 |
401.5 |
45.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.845 ± 0.015 | 1.973 ± 0.01 |
5.308 ± 0.012 | 6.916 ± 0.023 |
4.776 ± 0.016 | 5.127 ± 0.016 |
2.298 ± 0.007 | 6.225 ± 0.014 |
6.615 ± 0.018 | 8.661 ± 0.019 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.636 ± 0.009 | 4.939 ± 0.012 |
4.889 ± 0.021 | 4.012 ± 0.014 |
5.337 ± 0.015 | 7.961 ± 0.021 |
5.893 ± 0.019 | 6.242 ± 0.011 |
1.126 ± 0.004 | 3.211 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
