
Cricket associated circular virus 1
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses; Volvovirus
Average proteome isoelectric point is 8.29
Get precalculated fractions of proteins
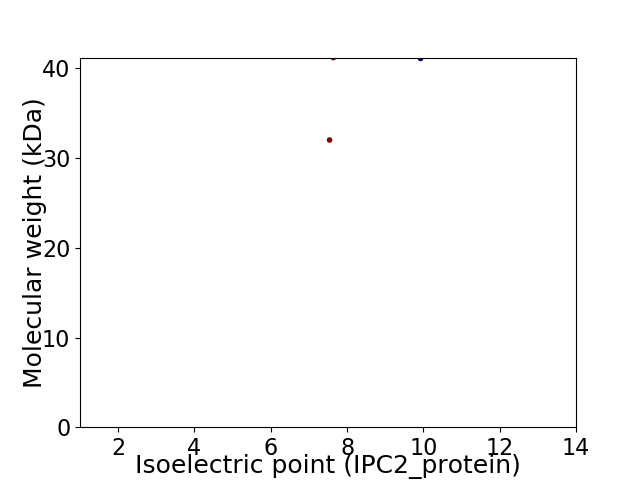
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346BPB7|A0A346BPB7_9VIRU Putative capsid protein OS=Cricket associated circular virus 1 OX=2293276 PE=4 SV=1
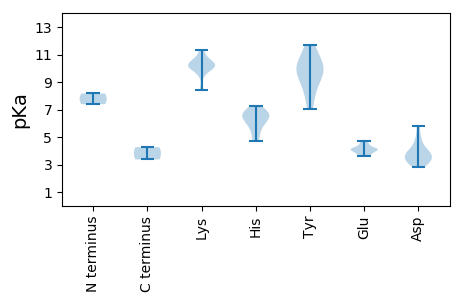
MM1 pKa = 8.17RR2 pKa = 11.84FRR4 pKa = 11.84HH5 pKa = 5.89WIGTSFNLEE14 pKa = 4.4SIPDD18 pKa = 3.87DD19 pKa = 3.66EE20 pKa = 4.71STIKK24 pKa = 10.55QIAYY28 pKa = 9.25QRR30 pKa = 11.84EE31 pKa = 4.13RR32 pKa = 11.84CPEE35 pKa = 3.89TNRR38 pKa = 11.84LHH40 pKa = 6.64LQFCISFTNPRR51 pKa = 11.84TMDD54 pKa = 3.12GVKK57 pKa = 9.97RR58 pKa = 11.84YY59 pKa = 10.08IGDD62 pKa = 3.37QSAHH66 pKa = 6.73LEE68 pKa = 3.89PCRR71 pKa = 11.84NLRR74 pKa = 11.84KK75 pKa = 10.06GLEE78 pKa = 4.07YY79 pKa = 10.75CNKK82 pKa = 10.09SEE84 pKa = 4.53SFVDD88 pKa = 3.4NRR90 pKa = 11.84FSRR93 pKa = 11.84FNTDD97 pKa = 2.85AQDD100 pKa = 3.69APDD103 pKa = 4.14DD104 pKa = 3.9FRR106 pKa = 11.84EE107 pKa = 4.26FTEE110 pKa = 4.05LQLWQKK116 pKa = 9.49YY117 pKa = 7.23PNWMLKK123 pKa = 10.13HH124 pKa = 5.18GTQVRR129 pKa = 11.84RR130 pKa = 11.84YY131 pKa = 8.23YY132 pKa = 9.92QICQEE137 pKa = 4.19VPQTRR142 pKa = 11.84EE143 pKa = 3.65KK144 pKa = 9.58PVCYY148 pKa = 10.02IFWGPAGTGKK158 pKa = 10.38SYY160 pKa = 10.84SARR163 pKa = 11.84HH164 pKa = 4.73WLGDD168 pKa = 3.31QLYY171 pKa = 9.31IKK173 pKa = 10.22PPGNFWIGYY182 pKa = 7.98NGEE185 pKa = 4.16KK186 pKa = 10.45AVLFDD191 pKa = 5.52DD192 pKa = 4.88YY193 pKa = 11.53YY194 pKa = 11.67SSEE197 pKa = 4.55KK198 pKa = 10.73YY199 pKa = 10.45DD200 pKa = 4.92DD201 pKa = 3.86LLRR204 pKa = 11.84WISEE208 pKa = 4.15NPIHH212 pKa = 7.27VSIKK216 pKa = 10.55GSSTPLKK223 pKa = 10.1AIKK226 pKa = 9.82FAFTSNMNPRR236 pKa = 11.84SWHH239 pKa = 5.88SKK241 pKa = 10.1IEE243 pKa = 3.95DD244 pKa = 3.16HH245 pKa = 6.79SALFRR250 pKa = 11.84RR251 pKa = 11.84ITKK254 pKa = 10.26CFFCTDD260 pKa = 2.95KK261 pKa = 11.23CFSLDD266 pKa = 3.7NLHH269 pKa = 6.6EE270 pKa = 4.26
MM1 pKa = 8.17RR2 pKa = 11.84FRR4 pKa = 11.84HH5 pKa = 5.89WIGTSFNLEE14 pKa = 4.4SIPDD18 pKa = 3.87DD19 pKa = 3.66EE20 pKa = 4.71STIKK24 pKa = 10.55QIAYY28 pKa = 9.25QRR30 pKa = 11.84EE31 pKa = 4.13RR32 pKa = 11.84CPEE35 pKa = 3.89TNRR38 pKa = 11.84LHH40 pKa = 6.64LQFCISFTNPRR51 pKa = 11.84TMDD54 pKa = 3.12GVKK57 pKa = 9.97RR58 pKa = 11.84YY59 pKa = 10.08IGDD62 pKa = 3.37QSAHH66 pKa = 6.73LEE68 pKa = 3.89PCRR71 pKa = 11.84NLRR74 pKa = 11.84KK75 pKa = 10.06GLEE78 pKa = 4.07YY79 pKa = 10.75CNKK82 pKa = 10.09SEE84 pKa = 4.53SFVDD88 pKa = 3.4NRR90 pKa = 11.84FSRR93 pKa = 11.84FNTDD97 pKa = 2.85AQDD100 pKa = 3.69APDD103 pKa = 4.14DD104 pKa = 3.9FRR106 pKa = 11.84EE107 pKa = 4.26FTEE110 pKa = 4.05LQLWQKK116 pKa = 9.49YY117 pKa = 7.23PNWMLKK123 pKa = 10.13HH124 pKa = 5.18GTQVRR129 pKa = 11.84RR130 pKa = 11.84YY131 pKa = 8.23YY132 pKa = 9.92QICQEE137 pKa = 4.19VPQTRR142 pKa = 11.84EE143 pKa = 3.65KK144 pKa = 9.58PVCYY148 pKa = 10.02IFWGPAGTGKK158 pKa = 10.38SYY160 pKa = 10.84SARR163 pKa = 11.84HH164 pKa = 4.73WLGDD168 pKa = 3.31QLYY171 pKa = 9.31IKK173 pKa = 10.22PPGNFWIGYY182 pKa = 7.98NGEE185 pKa = 4.16KK186 pKa = 10.45AVLFDD191 pKa = 5.52DD192 pKa = 4.88YY193 pKa = 11.53YY194 pKa = 11.67SSEE197 pKa = 4.55KK198 pKa = 10.73YY199 pKa = 10.45DD200 pKa = 4.92DD201 pKa = 3.86LLRR204 pKa = 11.84WISEE208 pKa = 4.15NPIHH212 pKa = 7.27VSIKK216 pKa = 10.55GSSTPLKK223 pKa = 10.1AIKK226 pKa = 9.82FAFTSNMNPRR236 pKa = 11.84SWHH239 pKa = 5.88SKK241 pKa = 10.1IEE243 pKa = 3.95DD244 pKa = 3.16HH245 pKa = 6.79SALFRR250 pKa = 11.84RR251 pKa = 11.84ITKK254 pKa = 10.26CFFCTDD260 pKa = 2.95KK261 pKa = 11.23CFSLDD266 pKa = 3.7NLHH269 pKa = 6.6EE270 pKa = 4.26
Molecular weight: 32.02 kDa
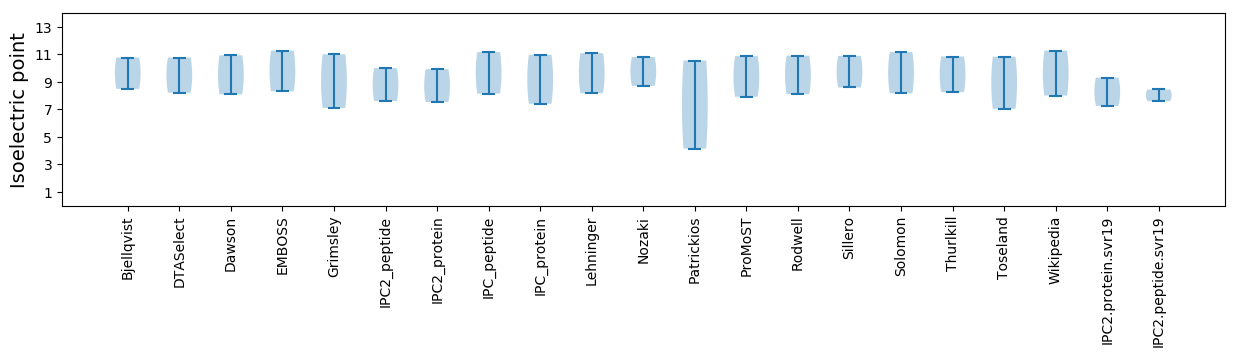
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346BPB7|A0A346BPB7_9VIRU Putative capsid protein OS=Cricket associated circular virus 1 OX=2293276 PE=4 SV=1
MM1 pKa = 7.39ARR3 pKa = 11.84LVKK6 pKa = 10.34SSRR9 pKa = 11.84RR10 pKa = 11.84LMHH13 pKa = 6.12RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84GFAIRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84YY27 pKa = 8.24SGRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84YY33 pKa = 9.93SSRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84YY39 pKa = 9.4SIRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84VSRR47 pKa = 11.84SLAIPRR53 pKa = 11.84RR54 pKa = 11.84GARR57 pKa = 11.84EE58 pKa = 3.79FSWVLNLHH66 pKa = 6.64NYY68 pKa = 7.06QHH70 pKa = 6.45SFNDD74 pKa = 3.55GGAISAIQGNQWYY87 pKa = 10.21LMSPPTLIIPYY98 pKa = 9.12QAFRR102 pKa = 11.84FLFKK106 pKa = 10.14EE107 pKa = 3.73WRR109 pKa = 11.84LTRR112 pKa = 11.84LLVAIRR118 pKa = 11.84RR119 pKa = 11.84RR120 pKa = 11.84YY121 pKa = 8.65PEE123 pKa = 3.84PVVYY127 pKa = 10.34QPNQQEE133 pKa = 4.08QTVAASRR140 pKa = 11.84LASNNIWWCPWGKK153 pKa = 10.26YY154 pKa = 9.25DD155 pKa = 5.83RR156 pKa = 11.84PEE158 pKa = 3.66QAPRR162 pKa = 11.84QTKK165 pKa = 10.02SAILLTSQWAVRR177 pKa = 11.84SVSQRR182 pKa = 11.84CQRR185 pKa = 11.84LSLVSTRR192 pKa = 11.84RR193 pKa = 11.84SASFVSGSTPIIDD206 pKa = 3.87GNEE209 pKa = 3.6GRR211 pKa = 11.84PGGGYY216 pKa = 9.91GQASLLSNVGFVAYY230 pKa = 9.27WGRR233 pKa = 11.84WPTYY237 pKa = 10.45SFSDD241 pKa = 3.98TYY243 pKa = 11.57NATPNSDD250 pKa = 4.1RR251 pKa = 11.84NDD253 pKa = 4.11LLPVANLGFIAVEE266 pKa = 4.05NLSEE270 pKa = 4.07ITPDD274 pKa = 3.16AVEE277 pKa = 4.22IKK279 pKa = 10.62VKK281 pKa = 10.88AFFSFKK287 pKa = 10.23GRR289 pKa = 11.84KK290 pKa = 8.57NLGSVNDD297 pKa = 3.65STGAFTSTFPLSSTDD312 pKa = 2.93WYY314 pKa = 11.07LDD316 pKa = 3.38NNKK319 pKa = 8.41TVPPVGVLEE328 pKa = 4.15QNLLDD333 pKa = 4.58QVDD336 pKa = 3.51KK337 pKa = 11.34DD338 pKa = 4.52EE339 pKa = 4.51PVEE342 pKa = 4.1MPDD345 pKa = 3.12IGPASVVAASPDD357 pKa = 3.33GPMVV361 pKa = 3.37
MM1 pKa = 7.39ARR3 pKa = 11.84LVKK6 pKa = 10.34SSRR9 pKa = 11.84RR10 pKa = 11.84LMHH13 pKa = 6.12RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84GFAIRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84YY27 pKa = 8.24SGRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84YY33 pKa = 9.93SSRR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84YY39 pKa = 9.4SIRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84VSRR47 pKa = 11.84SLAIPRR53 pKa = 11.84RR54 pKa = 11.84GARR57 pKa = 11.84EE58 pKa = 3.79FSWVLNLHH66 pKa = 6.64NYY68 pKa = 7.06QHH70 pKa = 6.45SFNDD74 pKa = 3.55GGAISAIQGNQWYY87 pKa = 10.21LMSPPTLIIPYY98 pKa = 9.12QAFRR102 pKa = 11.84FLFKK106 pKa = 10.14EE107 pKa = 3.73WRR109 pKa = 11.84LTRR112 pKa = 11.84LLVAIRR118 pKa = 11.84RR119 pKa = 11.84RR120 pKa = 11.84YY121 pKa = 8.65PEE123 pKa = 3.84PVVYY127 pKa = 10.34QPNQQEE133 pKa = 4.08QTVAASRR140 pKa = 11.84LASNNIWWCPWGKK153 pKa = 10.26YY154 pKa = 9.25DD155 pKa = 5.83RR156 pKa = 11.84PEE158 pKa = 3.66QAPRR162 pKa = 11.84QTKK165 pKa = 10.02SAILLTSQWAVRR177 pKa = 11.84SVSQRR182 pKa = 11.84CQRR185 pKa = 11.84LSLVSTRR192 pKa = 11.84RR193 pKa = 11.84SASFVSGSTPIIDD206 pKa = 3.87GNEE209 pKa = 3.6GRR211 pKa = 11.84PGGGYY216 pKa = 9.91GQASLLSNVGFVAYY230 pKa = 9.27WGRR233 pKa = 11.84WPTYY237 pKa = 10.45SFSDD241 pKa = 3.98TYY243 pKa = 11.57NATPNSDD250 pKa = 4.1RR251 pKa = 11.84NDD253 pKa = 4.11LLPVANLGFIAVEE266 pKa = 4.05NLSEE270 pKa = 4.07ITPDD274 pKa = 3.16AVEE277 pKa = 4.22IKK279 pKa = 10.62VKK281 pKa = 10.88AFFSFKK287 pKa = 10.23GRR289 pKa = 11.84KK290 pKa = 8.57NLGSVNDD297 pKa = 3.65STGAFTSTFPLSSTDD312 pKa = 2.93WYY314 pKa = 11.07LDD316 pKa = 3.38NNKK319 pKa = 8.41TVPPVGVLEE328 pKa = 4.15QNLLDD333 pKa = 4.58QVDD336 pKa = 3.51KK337 pKa = 11.34DD338 pKa = 4.52EE339 pKa = 4.51PVEE342 pKa = 4.1MPDD345 pKa = 3.12IGPASVVAASPDD357 pKa = 3.33GPMVV361 pKa = 3.37
Molecular weight: 41.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
631 |
270 |
361 |
315.5 |
36.57 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.705 ± 1.129 | 1.743 ± 0.897 |
5.071 ± 0.691 | 4.437 ± 0.84 |
5.23 ± 0.811 | 5.705 ± 0.502 |
1.902 ± 0.808 | 5.071 ± 0.482 |
4.279 ± 1.138 | 7.29 ± 0.352 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.426 ± 0.031 | 5.23 ± 0.025 |
6.022 ± 0.472 | 4.437 ± 0.004 |
9.984 ± 1.454 | 9.35 ± 0.887 |
4.754 ± 0.243 | 5.23 ± 1.488 |
2.853 ± 0.062 | 4.279 ± 0.302 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
