
Actinorugispora endophytica
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptosporangiales; Nocardiopsaceae; Actinorugispora
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
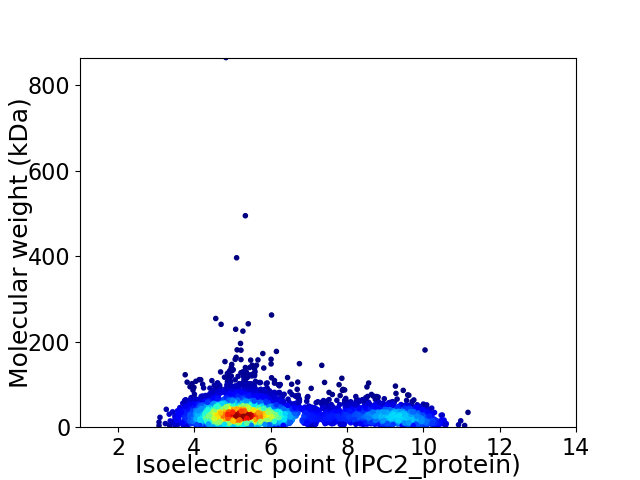
Virtual 2D-PAGE plot for 4593 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R6UL49|A0A4R6UL49_9ACTN Putative repeat protein (TIGR03847 family) OS=Actinorugispora endophytica OX=1605990 GN=EV190_12812 PE=4 SV=1
MM1 pKa = 7.34GFVALGAAATLLLSACGGGDD21 pKa = 3.6GGDD24 pKa = 3.57GDD26 pKa = 4.54GGSGAGGVFDD36 pKa = 3.73QASTEE41 pKa = 4.2VVNASDD47 pKa = 3.49ATGGTLRR54 pKa = 11.84YY55 pKa = 9.7AISANMEE62 pKa = 3.94STDD65 pKa = 4.47PGNTYY70 pKa = 10.58YY71 pKa = 11.08GYY73 pKa = 9.13VWNFSRR79 pKa = 11.84YY80 pKa = 6.71YY81 pKa = 11.04ARR83 pKa = 11.84TLLTFSPVPGQASSEE98 pKa = 4.08LLPDD102 pKa = 4.03LAEE105 pKa = 4.86DD106 pKa = 3.79MPEE109 pKa = 3.93VSDD112 pKa = 6.33DD113 pKa = 3.63GTEE116 pKa = 3.76WTIKK120 pKa = 10.28LKK122 pKa = 10.77QGLKK126 pKa = 10.39YY127 pKa = 10.34EE128 pKa = 4.75DD129 pKa = 3.73GSEE132 pKa = 4.19IVAEE136 pKa = 4.51DD137 pKa = 3.01IKK139 pKa = 11.38YY140 pKa = 10.72AIARR144 pKa = 11.84SNFGPQALPNGPKK157 pKa = 10.22YY158 pKa = 10.24FQQLLDD164 pKa = 3.96ADD166 pKa = 4.9DD167 pKa = 4.42YY168 pKa = 11.04EE169 pKa = 5.36GPYY172 pKa = 10.76GGGDD176 pKa = 3.52PLAGFDD182 pKa = 5.43AIEE185 pKa = 4.39TPDD188 pKa = 4.63DD189 pKa = 3.48YY190 pKa = 11.97TLVFKK195 pKa = 10.96LKK197 pKa = 10.8APFADD202 pKa = 4.51FPYY205 pKa = 10.37ILSQTQTAPVPAEE218 pKa = 3.99ADD220 pKa = 2.87TGEE223 pKa = 4.17QYY225 pKa = 10.57QGQVVSSGPYY235 pKa = 9.44KK236 pKa = 10.75FEE238 pKa = 3.99GSYY241 pKa = 10.3TPGDD245 pKa = 3.52GLVLVRR251 pKa = 11.84NDD253 pKa = 2.92QWDD256 pKa = 3.93PEE258 pKa = 4.28SDD260 pKa = 3.51PTRR263 pKa = 11.84EE264 pKa = 4.03ALPDD268 pKa = 3.27RR269 pKa = 11.84VTVEE273 pKa = 3.91EE274 pKa = 4.42GVEE277 pKa = 3.9QNEE280 pKa = 3.65IDD282 pKa = 3.49QRR284 pKa = 11.84LVNGEE289 pKa = 4.53LDD291 pKa = 3.29VDD293 pKa = 4.44LAGTGLGPAMKK304 pKa = 10.3GDD306 pKa = 4.25LLADD310 pKa = 3.57EE311 pKa = 4.71EE312 pKa = 4.35ARR314 pKa = 11.84ANIDD318 pKa = 3.46NPVTGAHH325 pKa = 6.65FYY327 pKa = 10.85VAINTKK333 pKa = 9.48VEE335 pKa = 4.01PLDD338 pKa = 5.41DD339 pKa = 3.91VACRR343 pKa = 11.84QAIQYY348 pKa = 8.02AASRR352 pKa = 11.84EE353 pKa = 4.48GIQRR357 pKa = 11.84AYY359 pKa = 10.2GGEE362 pKa = 4.04FGGEE366 pKa = 3.66IATQVLPPSIPGANPEE382 pKa = 4.2LDD384 pKa = 3.77PYY386 pKa = 10.22EE387 pKa = 4.41AASGTGNVEE396 pKa = 3.68MAQEE400 pKa = 3.94KK401 pKa = 10.81LEE403 pKa = 4.23EE404 pKa = 4.6CGEE407 pKa = 4.1EE408 pKa = 5.01DD409 pKa = 4.48GFTVNIGVRR418 pKa = 11.84SDD420 pKa = 3.44RR421 pKa = 11.84PAEE424 pKa = 4.01VAAVEE429 pKa = 4.67SIQEE433 pKa = 4.0SLAAANITAEE443 pKa = 4.27IKK445 pKa = 10.66SFPSDD450 pKa = 3.01TFTNTQAGSPDD461 pKa = 3.4FVADD465 pKa = 3.7NEE467 pKa = 4.68LGLNYY472 pKa = 9.67YY473 pKa = 10.48GWMSDD478 pKa = 2.85WPSGYY483 pKa = 10.84GYY485 pKa = 9.92MSSILDD491 pKa = 3.46GDD493 pKa = 4.46SIKK496 pKa = 10.82EE497 pKa = 3.95AGNSNISEE505 pKa = 4.32LDD507 pKa = 3.48DD508 pKa = 4.21PEE510 pKa = 5.09INQLFDD516 pKa = 4.94DD517 pKa = 4.31VTQVEE522 pKa = 4.63DD523 pKa = 4.38PEE525 pKa = 4.18EE526 pKa = 3.63QAAIYY531 pKa = 9.78SQIDD535 pKa = 3.85EE536 pKa = 4.65LTMEE540 pKa = 4.77SATILPGVFQKK551 pKa = 10.91SVLYY555 pKa = 10.47RR556 pKa = 11.84PEE558 pKa = 4.05NLTNVYY564 pKa = 9.83FNPSYY569 pKa = 11.38HH570 pKa = 6.5MYY572 pKa = 10.8DD573 pKa = 3.56YY574 pKa = 10.29MALGTTNNEE583 pKa = 3.79
MM1 pKa = 7.34GFVALGAAATLLLSACGGGDD21 pKa = 3.6GGDD24 pKa = 3.57GDD26 pKa = 4.54GGSGAGGVFDD36 pKa = 3.73QASTEE41 pKa = 4.2VVNASDD47 pKa = 3.49ATGGTLRR54 pKa = 11.84YY55 pKa = 9.7AISANMEE62 pKa = 3.94STDD65 pKa = 4.47PGNTYY70 pKa = 10.58YY71 pKa = 11.08GYY73 pKa = 9.13VWNFSRR79 pKa = 11.84YY80 pKa = 6.71YY81 pKa = 11.04ARR83 pKa = 11.84TLLTFSPVPGQASSEE98 pKa = 4.08LLPDD102 pKa = 4.03LAEE105 pKa = 4.86DD106 pKa = 3.79MPEE109 pKa = 3.93VSDD112 pKa = 6.33DD113 pKa = 3.63GTEE116 pKa = 3.76WTIKK120 pKa = 10.28LKK122 pKa = 10.77QGLKK126 pKa = 10.39YY127 pKa = 10.34EE128 pKa = 4.75DD129 pKa = 3.73GSEE132 pKa = 4.19IVAEE136 pKa = 4.51DD137 pKa = 3.01IKK139 pKa = 11.38YY140 pKa = 10.72AIARR144 pKa = 11.84SNFGPQALPNGPKK157 pKa = 10.22YY158 pKa = 10.24FQQLLDD164 pKa = 3.96ADD166 pKa = 4.9DD167 pKa = 4.42YY168 pKa = 11.04EE169 pKa = 5.36GPYY172 pKa = 10.76GGGDD176 pKa = 3.52PLAGFDD182 pKa = 5.43AIEE185 pKa = 4.39TPDD188 pKa = 4.63DD189 pKa = 3.48YY190 pKa = 11.97TLVFKK195 pKa = 10.96LKK197 pKa = 10.8APFADD202 pKa = 4.51FPYY205 pKa = 10.37ILSQTQTAPVPAEE218 pKa = 3.99ADD220 pKa = 2.87TGEE223 pKa = 4.17QYY225 pKa = 10.57QGQVVSSGPYY235 pKa = 9.44KK236 pKa = 10.75FEE238 pKa = 3.99GSYY241 pKa = 10.3TPGDD245 pKa = 3.52GLVLVRR251 pKa = 11.84NDD253 pKa = 2.92QWDD256 pKa = 3.93PEE258 pKa = 4.28SDD260 pKa = 3.51PTRR263 pKa = 11.84EE264 pKa = 4.03ALPDD268 pKa = 3.27RR269 pKa = 11.84VTVEE273 pKa = 3.91EE274 pKa = 4.42GVEE277 pKa = 3.9QNEE280 pKa = 3.65IDD282 pKa = 3.49QRR284 pKa = 11.84LVNGEE289 pKa = 4.53LDD291 pKa = 3.29VDD293 pKa = 4.44LAGTGLGPAMKK304 pKa = 10.3GDD306 pKa = 4.25LLADD310 pKa = 3.57EE311 pKa = 4.71EE312 pKa = 4.35ARR314 pKa = 11.84ANIDD318 pKa = 3.46NPVTGAHH325 pKa = 6.65FYY327 pKa = 10.85VAINTKK333 pKa = 9.48VEE335 pKa = 4.01PLDD338 pKa = 5.41DD339 pKa = 3.91VACRR343 pKa = 11.84QAIQYY348 pKa = 8.02AASRR352 pKa = 11.84EE353 pKa = 4.48GIQRR357 pKa = 11.84AYY359 pKa = 10.2GGEE362 pKa = 4.04FGGEE366 pKa = 3.66IATQVLPPSIPGANPEE382 pKa = 4.2LDD384 pKa = 3.77PYY386 pKa = 10.22EE387 pKa = 4.41AASGTGNVEE396 pKa = 3.68MAQEE400 pKa = 3.94KK401 pKa = 10.81LEE403 pKa = 4.23EE404 pKa = 4.6CGEE407 pKa = 4.1EE408 pKa = 5.01DD409 pKa = 4.48GFTVNIGVRR418 pKa = 11.84SDD420 pKa = 3.44RR421 pKa = 11.84PAEE424 pKa = 4.01VAAVEE429 pKa = 4.67SIQEE433 pKa = 4.0SLAAANITAEE443 pKa = 4.27IKK445 pKa = 10.66SFPSDD450 pKa = 3.01TFTNTQAGSPDD461 pKa = 3.4FVADD465 pKa = 3.7NEE467 pKa = 4.68LGLNYY472 pKa = 9.67YY473 pKa = 10.48GWMSDD478 pKa = 2.85WPSGYY483 pKa = 10.84GYY485 pKa = 9.92MSSILDD491 pKa = 3.46GDD493 pKa = 4.46SIKK496 pKa = 10.82EE497 pKa = 3.95AGNSNISEE505 pKa = 4.32LDD507 pKa = 3.48DD508 pKa = 4.21PEE510 pKa = 5.09INQLFDD516 pKa = 4.94DD517 pKa = 4.31VTQVEE522 pKa = 4.63DD523 pKa = 4.38PEE525 pKa = 4.18EE526 pKa = 3.63QAAIYY531 pKa = 9.78SQIDD535 pKa = 3.85EE536 pKa = 4.65LTMEE540 pKa = 4.77SATILPGVFQKK551 pKa = 10.91SVLYY555 pKa = 10.47RR556 pKa = 11.84PEE558 pKa = 4.05NLTNVYY564 pKa = 9.83FNPSYY569 pKa = 11.38HH570 pKa = 6.5MYY572 pKa = 10.8DD573 pKa = 3.56YY574 pKa = 10.29MALGTTNNEE583 pKa = 3.79
Molecular weight: 62.55 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R6UNQ8|A0A4R6UNQ8_9ACTN Cytochrome P450 OS=Actinorugispora endophytica OX=1605990 GN=EV190_11744 PE=3 SV=1
MM1 pKa = 6.35MTGAAATAAPTARR14 pKa = 11.84AVSAAGPPRR23 pKa = 11.84AVSAANPTSPGAATATATSTARR45 pKa = 11.84AAPTGAVGTTVTRR58 pKa = 11.84VVAVPTAVTVTTARR72 pKa = 11.84VAGTATVTRR81 pKa = 11.84AVAVPTAVTVTTARR95 pKa = 11.84VAGTATVTRR104 pKa = 11.84AVAPVVATTGGAHH117 pKa = 6.1VAPATATTVPVAGTATATRR136 pKa = 11.84AVARR140 pKa = 11.84VVATTARR147 pKa = 11.84AAVPLGAVGMTVLVVMIGVARR168 pKa = 11.84VRR170 pKa = 11.84AVRR173 pKa = 11.84SSAATTARR181 pKa = 11.84VAGTATATRR190 pKa = 11.84VAAVPTAVTVTTARR204 pKa = 11.84VAGTATATRR213 pKa = 11.84VVARR217 pKa = 11.84VRR219 pKa = 11.84AVRR222 pKa = 11.84SNVVTTVPVAGTATVTRR239 pKa = 11.84AVARR243 pKa = 11.84VVATTVRR250 pKa = 11.84AAVPLGAVGMTVLVVMIGVARR271 pKa = 11.84VRR273 pKa = 11.84AVRR276 pKa = 11.84SSAATTAPAAVPPVAGPPSARR297 pKa = 11.84AAPSATCRR305 pKa = 11.84PRR307 pKa = 11.84PRR309 pKa = 11.84PGSAPFARR317 pKa = 11.84STTPATTTRR326 pKa = 11.84PTTRR330 pKa = 11.84VTATSTCPAASGCRR344 pKa = 11.84RR345 pKa = 11.84PSPRR349 pKa = 11.84RR350 pKa = 11.84VWPAGAPARR359 pKa = 11.84SS360 pKa = 3.37
MM1 pKa = 6.35MTGAAATAAPTARR14 pKa = 11.84AVSAAGPPRR23 pKa = 11.84AVSAANPTSPGAATATATSTARR45 pKa = 11.84AAPTGAVGTTVTRR58 pKa = 11.84VVAVPTAVTVTTARR72 pKa = 11.84VAGTATVTRR81 pKa = 11.84AVAVPTAVTVTTARR95 pKa = 11.84VAGTATVTRR104 pKa = 11.84AVAPVVATTGGAHH117 pKa = 6.1VAPATATTVPVAGTATATRR136 pKa = 11.84AVARR140 pKa = 11.84VVATTARR147 pKa = 11.84AAVPLGAVGMTVLVVMIGVARR168 pKa = 11.84VRR170 pKa = 11.84AVRR173 pKa = 11.84SSAATTARR181 pKa = 11.84VAGTATATRR190 pKa = 11.84VAAVPTAVTVTTARR204 pKa = 11.84VAGTATATRR213 pKa = 11.84VVARR217 pKa = 11.84VRR219 pKa = 11.84AVRR222 pKa = 11.84SNVVTTVPVAGTATVTRR239 pKa = 11.84AVARR243 pKa = 11.84VVATTVRR250 pKa = 11.84AAVPLGAVGMTVLVVMIGVARR271 pKa = 11.84VRR273 pKa = 11.84AVRR276 pKa = 11.84SSAATTAPAAVPPVAGPPSARR297 pKa = 11.84AAPSATCRR305 pKa = 11.84PRR307 pKa = 11.84PRR309 pKa = 11.84PGSAPFARR317 pKa = 11.84STTPATTTRR326 pKa = 11.84PTTRR330 pKa = 11.84VTATSTCPAASGCRR344 pKa = 11.84RR345 pKa = 11.84PSPRR349 pKa = 11.84RR350 pKa = 11.84VWPAGAPARR359 pKa = 11.84SS360 pKa = 3.37
Molecular weight: 34.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1481852 |
29 |
7981 |
322.6 |
34.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.476 ± 0.057 | 0.715 ± 0.012 |
6.237 ± 0.038 | 6.29 ± 0.04 |
2.767 ± 0.019 | 9.654 ± 0.037 |
2.127 ± 0.018 | 3.042 ± 0.029 |
1.579 ± 0.024 | 10.361 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.765 ± 0.017 | 1.739 ± 0.018 |
6.195 ± 0.046 | 2.349 ± 0.025 |
8.658 ± 0.038 | 5.091 ± 0.028 |
5.532 ± 0.028 | 8.96 ± 0.044 |
1.465 ± 0.015 | 1.997 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
