
Bacteroides uniformis CAG:3
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; environmental samples
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
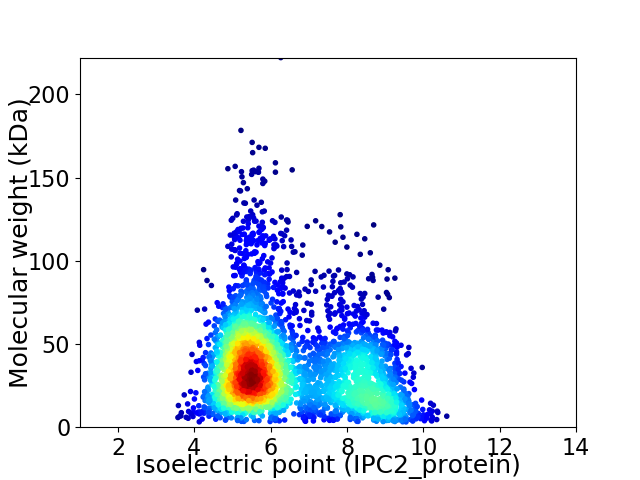
Virtual 2D-PAGE plot for 3570 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R7ELP5|R7ELP5_9BACE Uncharacterized protein OS=Bacteroides uniformis CAG:3 OX=1263055 GN=BN594_03137 PE=4 SV=1
MM1 pKa = 7.66KK2 pKa = 9.42MFKK5 pKa = 10.12MMAASMLGVALCLGFTACSDD25 pKa = 3.65DD26 pKa = 4.64DD27 pKa = 4.19EE28 pKa = 5.89NEE30 pKa = 4.03NGEE33 pKa = 4.23GGEE36 pKa = 4.17NTATVVNPSQVFTGGLPKK54 pKa = 10.24SVSGMAISHH63 pKa = 5.76NEE65 pKa = 3.67EE66 pKa = 3.83GLVTNITTEE75 pKa = 4.47DD76 pKa = 3.28GDD78 pKa = 4.01KK79 pKa = 11.19AVFEE83 pKa = 4.47YY84 pKa = 10.84FPATTKK90 pKa = 11.14ADD92 pKa = 3.59VAKK95 pKa = 10.44DD96 pKa = 3.37RR97 pKa = 11.84ARR99 pKa = 11.84ITVTDD104 pKa = 3.57EE105 pKa = 4.11EE106 pKa = 5.85GDD108 pKa = 3.78VTEE111 pKa = 6.43LNLQLNSDD119 pKa = 4.49GYY121 pKa = 11.63VEE123 pKa = 4.55FCNSIDD129 pKa = 4.12HH130 pKa = 7.04AGTPDD135 pKa = 3.45ADD137 pKa = 3.57EE138 pKa = 4.77FTWEE142 pKa = 4.13MEE144 pKa = 3.98YY145 pKa = 8.99DD146 pKa = 3.89TEE148 pKa = 4.14GHH150 pKa = 5.9LVVMKK155 pKa = 10.47RR156 pKa = 11.84SEE158 pKa = 4.23SDD160 pKa = 3.69GEE162 pKa = 4.25ITNITYY168 pKa = 10.65KK169 pKa = 10.94DD170 pKa = 3.27GDD172 pKa = 3.97VVKK175 pKa = 10.15TSTRR179 pKa = 11.84YY180 pKa = 9.26VASGDD185 pKa = 3.82LNGDD189 pKa = 4.18GIIDD193 pKa = 4.59SNDD196 pKa = 2.66EE197 pKa = 4.02WEE199 pKa = 4.23YY200 pKa = 11.45SAAIDD205 pKa = 3.85YY206 pKa = 7.7TTDD209 pKa = 3.53NITAPIEE216 pKa = 4.16NKK218 pKa = 9.95GCLMLFDD225 pKa = 5.86EE226 pKa = 5.11ILDD229 pKa = 3.59VDD231 pKa = 4.43MDD233 pKa = 3.88EE234 pKa = 5.25MIYY237 pKa = 10.46AYY239 pKa = 10.42YY240 pKa = 11.07GGMLGKK246 pKa = 8.84ATKK249 pKa = 9.95HH250 pKa = 5.63LPLVGHH256 pKa = 5.31YY257 pKa = 8.47TYY259 pKa = 11.0NGEE262 pKa = 4.28DD263 pKa = 3.85SVSDD267 pKa = 3.74MYY269 pKa = 9.7FTWTLNSDD277 pKa = 4.1SYY279 pKa = 10.13PTEE282 pKa = 4.38LVVKK286 pKa = 8.81DD287 pKa = 3.09QWDD290 pKa = 3.81EE291 pKa = 3.98YY292 pKa = 11.04RR293 pKa = 11.84CTFTWW298 pKa = 3.12
MM1 pKa = 7.66KK2 pKa = 9.42MFKK5 pKa = 10.12MMAASMLGVALCLGFTACSDD25 pKa = 3.65DD26 pKa = 4.64DD27 pKa = 4.19EE28 pKa = 5.89NEE30 pKa = 4.03NGEE33 pKa = 4.23GGEE36 pKa = 4.17NTATVVNPSQVFTGGLPKK54 pKa = 10.24SVSGMAISHH63 pKa = 5.76NEE65 pKa = 3.67EE66 pKa = 3.83GLVTNITTEE75 pKa = 4.47DD76 pKa = 3.28GDD78 pKa = 4.01KK79 pKa = 11.19AVFEE83 pKa = 4.47YY84 pKa = 10.84FPATTKK90 pKa = 11.14ADD92 pKa = 3.59VAKK95 pKa = 10.44DD96 pKa = 3.37RR97 pKa = 11.84ARR99 pKa = 11.84ITVTDD104 pKa = 3.57EE105 pKa = 4.11EE106 pKa = 5.85GDD108 pKa = 3.78VTEE111 pKa = 6.43LNLQLNSDD119 pKa = 4.49GYY121 pKa = 11.63VEE123 pKa = 4.55FCNSIDD129 pKa = 4.12HH130 pKa = 7.04AGTPDD135 pKa = 3.45ADD137 pKa = 3.57EE138 pKa = 4.77FTWEE142 pKa = 4.13MEE144 pKa = 3.98YY145 pKa = 8.99DD146 pKa = 3.89TEE148 pKa = 4.14GHH150 pKa = 5.9LVVMKK155 pKa = 10.47RR156 pKa = 11.84SEE158 pKa = 4.23SDD160 pKa = 3.69GEE162 pKa = 4.25ITNITYY168 pKa = 10.65KK169 pKa = 10.94DD170 pKa = 3.27GDD172 pKa = 3.97VVKK175 pKa = 10.15TSTRR179 pKa = 11.84YY180 pKa = 9.26VASGDD185 pKa = 3.82LNGDD189 pKa = 4.18GIIDD193 pKa = 4.59SNDD196 pKa = 2.66EE197 pKa = 4.02WEE199 pKa = 4.23YY200 pKa = 11.45SAAIDD205 pKa = 3.85YY206 pKa = 7.7TTDD209 pKa = 3.53NITAPIEE216 pKa = 4.16NKK218 pKa = 9.95GCLMLFDD225 pKa = 5.86EE226 pKa = 5.11ILDD229 pKa = 3.59VDD231 pKa = 4.43MDD233 pKa = 3.88EE234 pKa = 5.25MIYY237 pKa = 10.46AYY239 pKa = 10.42YY240 pKa = 11.07GGMLGKK246 pKa = 8.84ATKK249 pKa = 9.95HH250 pKa = 5.63LPLVGHH256 pKa = 5.31YY257 pKa = 8.47TYY259 pKa = 11.0NGEE262 pKa = 4.28DD263 pKa = 3.85SVSDD267 pKa = 3.74MYY269 pKa = 9.7FTWTLNSDD277 pKa = 4.1SYY279 pKa = 10.13PTEE282 pKa = 4.38LVVKK286 pKa = 8.81DD287 pKa = 3.09QWDD290 pKa = 3.81EE291 pKa = 3.98YY292 pKa = 11.04RR293 pKa = 11.84CTFTWW298 pKa = 3.12
Molecular weight: 33.02 kDa
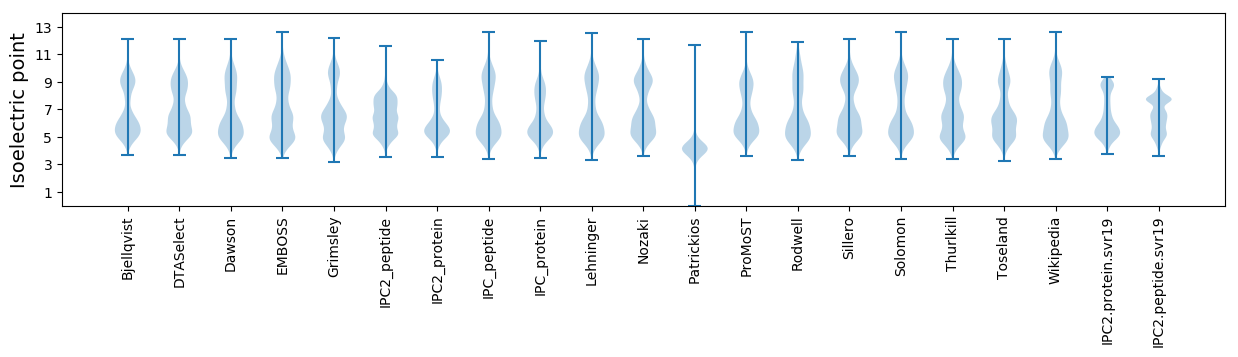
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R7EGQ5|R7EGQ5_9BACE Uncharacterized protein OS=Bacteroides uniformis CAG:3 OX=1263055 GN=BN594_02515 PE=4 SV=1
MM1 pKa = 7.63DD2 pKa = 4.52HH3 pKa = 6.94AAVIRR8 pKa = 11.84RR9 pKa = 11.84TLFGRR14 pKa = 11.84NRR16 pKa = 11.84RR17 pKa = 11.84QLRR20 pKa = 11.84IFGGRR25 pKa = 11.84HH26 pKa = 4.13KK27 pKa = 10.84FKK29 pKa = 10.21IYY31 pKa = 10.55RR32 pKa = 11.84MAVAGSLIHH41 pKa = 6.41LHH43 pKa = 5.23NQFMVLTEE51 pKa = 4.04RR52 pKa = 11.84QFGIAEE58 pKa = 4.1FSVFGSPYY66 pKa = 9.75QHH68 pKa = 5.69VTLTGRR74 pKa = 11.84QKK76 pKa = 10.98SLSLCIGLLPGYY88 pKa = 9.81SIEE91 pKa = 3.9LRR93 pKa = 11.84IIIEE97 pKa = 4.39LEE99 pKa = 3.66LHH101 pKa = 5.19TSIRR105 pKa = 11.84HH106 pKa = 5.6RR107 pKa = 11.84ISMSVHH113 pKa = 6.33DD114 pKa = 5.03RR115 pKa = 11.84NCGLSRR121 pKa = 11.84RR122 pKa = 11.84CIVTDD127 pKa = 3.87YY128 pKa = 11.1IDD130 pKa = 3.77FRR132 pKa = 11.84ISRR135 pKa = 11.84SDD137 pKa = 2.74RR138 pKa = 11.84HH139 pKa = 5.61HH140 pKa = 6.84FLRR143 pKa = 11.84SFITSEE149 pKa = 3.9DD150 pKa = 3.52LGMHH154 pKa = 5.28QHH156 pKa = 6.04TATGRR161 pKa = 11.84SIEE164 pKa = 4.07PTKK167 pKa = 10.62VEE169 pKa = 3.97YY170 pKa = 10.72RR171 pKa = 11.84FGFAGSKK178 pKa = 10.02KK179 pKa = 10.07IPFPVCPCFYY189 pKa = 10.32PRR191 pKa = 11.84MIVVGMRR198 pKa = 11.84PPGCVDD204 pKa = 3.22LTGRR208 pKa = 11.84DD209 pKa = 3.31THH211 pKa = 6.08RR212 pKa = 11.84TQGGNQQGRR221 pKa = 11.84FLATTPVCRR230 pKa = 11.84AYY232 pKa = 10.38RR233 pKa = 11.84CQRR236 pKa = 11.84RR237 pKa = 11.84TGTGIGGSIDD247 pKa = 3.83SLLVAPVVHH256 pKa = 6.49LQNGIIEE263 pKa = 4.36GQRR266 pKa = 11.84LDD268 pKa = 3.87TILQFFVKK276 pKa = 10.07HH277 pKa = 5.19YY278 pKa = 8.63PCTVQMLIVDD288 pKa = 3.71PHH290 pKa = 6.98RR291 pKa = 11.84KK292 pKa = 9.61YY293 pKa = 11.49KK294 pKa = 7.73MTKK297 pKa = 9.59QILRR301 pKa = 11.84NNFSPRR307 pKa = 11.84HH308 pKa = 5.59LIGSLLGGSHH318 pKa = 5.59IHH320 pKa = 5.49QMKK323 pKa = 10.07FSGIIGNVCQRR334 pKa = 11.84HH335 pKa = 5.27IGIKK339 pKa = 9.49EE340 pKa = 3.89LQSFAFVRR348 pKa = 11.84RR349 pKa = 11.84QVHH352 pKa = 5.6IEE354 pKa = 3.83YY355 pKa = 10.19GKK357 pKa = 10.6QIALRR362 pKa = 11.84QPFFFLIEE370 pKa = 4.24VLLHH374 pKa = 5.55TLTVRR379 pKa = 11.84SVCNEE384 pKa = 3.51RR385 pKa = 11.84TFAARR390 pKa = 11.84GKK392 pKa = 8.11QTYY395 pKa = 8.76QSEE398 pKa = 4.54KK399 pKa = 9.95SQKK402 pKa = 9.85SCNSFHH408 pKa = 6.3NAIIKK413 pKa = 9.4VEE415 pKa = 3.98PQKK418 pKa = 11.41
MM1 pKa = 7.63DD2 pKa = 4.52HH3 pKa = 6.94AAVIRR8 pKa = 11.84RR9 pKa = 11.84TLFGRR14 pKa = 11.84NRR16 pKa = 11.84RR17 pKa = 11.84QLRR20 pKa = 11.84IFGGRR25 pKa = 11.84HH26 pKa = 4.13KK27 pKa = 10.84FKK29 pKa = 10.21IYY31 pKa = 10.55RR32 pKa = 11.84MAVAGSLIHH41 pKa = 6.41LHH43 pKa = 5.23NQFMVLTEE51 pKa = 4.04RR52 pKa = 11.84QFGIAEE58 pKa = 4.1FSVFGSPYY66 pKa = 9.75QHH68 pKa = 5.69VTLTGRR74 pKa = 11.84QKK76 pKa = 10.98SLSLCIGLLPGYY88 pKa = 9.81SIEE91 pKa = 3.9LRR93 pKa = 11.84IIIEE97 pKa = 4.39LEE99 pKa = 3.66LHH101 pKa = 5.19TSIRR105 pKa = 11.84HH106 pKa = 5.6RR107 pKa = 11.84ISMSVHH113 pKa = 6.33DD114 pKa = 5.03RR115 pKa = 11.84NCGLSRR121 pKa = 11.84RR122 pKa = 11.84CIVTDD127 pKa = 3.87YY128 pKa = 11.1IDD130 pKa = 3.77FRR132 pKa = 11.84ISRR135 pKa = 11.84SDD137 pKa = 2.74RR138 pKa = 11.84HH139 pKa = 5.61HH140 pKa = 6.84FLRR143 pKa = 11.84SFITSEE149 pKa = 3.9DD150 pKa = 3.52LGMHH154 pKa = 5.28QHH156 pKa = 6.04TATGRR161 pKa = 11.84SIEE164 pKa = 4.07PTKK167 pKa = 10.62VEE169 pKa = 3.97YY170 pKa = 10.72RR171 pKa = 11.84FGFAGSKK178 pKa = 10.02KK179 pKa = 10.07IPFPVCPCFYY189 pKa = 10.32PRR191 pKa = 11.84MIVVGMRR198 pKa = 11.84PPGCVDD204 pKa = 3.22LTGRR208 pKa = 11.84DD209 pKa = 3.31THH211 pKa = 6.08RR212 pKa = 11.84TQGGNQQGRR221 pKa = 11.84FLATTPVCRR230 pKa = 11.84AYY232 pKa = 10.38RR233 pKa = 11.84CQRR236 pKa = 11.84RR237 pKa = 11.84TGTGIGGSIDD247 pKa = 3.83SLLVAPVVHH256 pKa = 6.49LQNGIIEE263 pKa = 4.36GQRR266 pKa = 11.84LDD268 pKa = 3.87TILQFFVKK276 pKa = 10.07HH277 pKa = 5.19YY278 pKa = 8.63PCTVQMLIVDD288 pKa = 3.71PHH290 pKa = 6.98RR291 pKa = 11.84KK292 pKa = 9.61YY293 pKa = 11.49KK294 pKa = 7.73MTKK297 pKa = 9.59QILRR301 pKa = 11.84NNFSPRR307 pKa = 11.84HH308 pKa = 5.59LIGSLLGGSHH318 pKa = 5.59IHH320 pKa = 5.49QMKK323 pKa = 10.07FSGIIGNVCQRR334 pKa = 11.84HH335 pKa = 5.27IGIKK339 pKa = 9.49EE340 pKa = 3.89LQSFAFVRR348 pKa = 11.84RR349 pKa = 11.84QVHH352 pKa = 5.6IEE354 pKa = 3.83YY355 pKa = 10.19GKK357 pKa = 10.6QIALRR362 pKa = 11.84QPFFFLIEE370 pKa = 4.24VLLHH374 pKa = 5.55TLTVRR379 pKa = 11.84SVCNEE384 pKa = 3.51RR385 pKa = 11.84TFAARR390 pKa = 11.84GKK392 pKa = 8.11QTYY395 pKa = 8.76QSEE398 pKa = 4.54KK399 pKa = 9.95SQKK402 pKa = 9.85SCNSFHH408 pKa = 6.3NAIIKK413 pKa = 9.4VEE415 pKa = 3.98PQKK418 pKa = 11.41
Molecular weight: 47.91 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1263016 |
29 |
1965 |
353.8 |
39.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.378 ± 0.043 | 1.29 ± 0.017 |
5.47 ± 0.032 | 6.578 ± 0.042 |
4.568 ± 0.028 | 6.997 ± 0.036 |
1.956 ± 0.016 | 6.639 ± 0.034 |
6.362 ± 0.037 | 9.185 ± 0.042 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.81 ± 0.017 | 4.929 ± 0.041 |
3.796 ± 0.023 | 3.452 ± 0.019 |
4.757 ± 0.03 | 5.972 ± 0.034 |
5.592 ± 0.032 | 6.528 ± 0.032 |
1.296 ± 0.016 | 4.445 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
