
Beihai barnacle virus 13
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.87
Get precalculated fractions of proteins
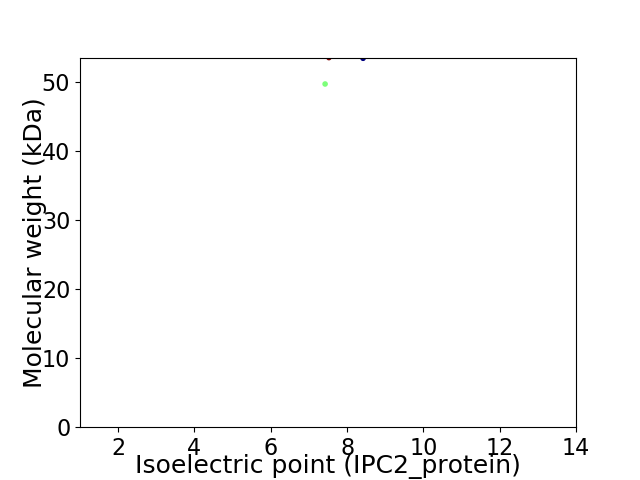
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KLI1|A0A1L3KLI1_9VIRU RdRp OS=Beihai barnacle virus 13 OX=1922357 PE=4 SV=1
MM1 pKa = 7.66DD2 pKa = 4.59TSTSQPSDD10 pKa = 3.44EE11 pKa = 4.79RR12 pKa = 11.84EE13 pKa = 3.92SAPTHH18 pKa = 5.79FRR20 pKa = 11.84SSSNNKK26 pKa = 9.09QNSTNQSGRR35 pKa = 11.84GGARR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84GRR43 pKa = 11.84ASSEE47 pKa = 3.66KK48 pKa = 10.32HH49 pKa = 5.04NEE51 pKa = 3.64AMRR54 pKa = 11.84KK55 pKa = 7.57FHH57 pKa = 7.2GPSQLQAYY65 pKa = 7.93RR66 pKa = 11.84AQLASNNFLDD76 pKa = 5.1LAEE79 pKa = 5.33LSQQLASLASASTPLTVKK97 pKa = 10.76LPATTRR103 pKa = 11.84GLAEE107 pKa = 4.17AVLGGYY113 pKa = 10.25HH114 pKa = 5.5MAFRR118 pKa = 11.84LTKK121 pKa = 10.13EE122 pKa = 4.07PPISLNAFYY131 pKa = 10.58RR132 pKa = 11.84VTLAQFEE139 pKa = 4.39LQAIRR144 pKa = 11.84SQLRR148 pKa = 11.84DD149 pKa = 3.41HH150 pKa = 6.86NISLRR155 pKa = 11.84QYY157 pKa = 9.62HH158 pKa = 5.45YY159 pKa = 11.03EE160 pKa = 3.95EE161 pKa = 5.15FFAKK165 pKa = 10.14YY166 pKa = 10.57ASFEE170 pKa = 4.39QIVAAHH176 pKa = 7.0PATLKK181 pKa = 10.93LVVAVIEE188 pKa = 4.25QYY190 pKa = 9.96GTVSVGGKK198 pKa = 7.82TYY200 pKa = 11.09SPFIVDD206 pKa = 4.9SNVDD210 pKa = 2.87WSVEE214 pKa = 3.52VRR216 pKa = 11.84ARR218 pKa = 11.84RR219 pKa = 11.84RR220 pKa = 11.84GHH222 pKa = 6.4RR223 pKa = 11.84ATGASTSALPHH234 pKa = 6.3RR235 pKa = 11.84VLEE238 pKa = 4.82DD239 pKa = 3.16GGEE242 pKa = 4.27RR243 pKa = 11.84ILEE246 pKa = 4.11VRR248 pKa = 11.84KK249 pKa = 10.62GMGPDD254 pKa = 3.55PYY256 pKa = 10.81TVTFRR261 pKa = 11.84NLEE264 pKa = 3.87NVVRR268 pKa = 11.84VMSDD272 pKa = 3.1EE273 pKa = 4.03NSVLEE278 pKa = 3.94GRR280 pKa = 11.84KK281 pKa = 8.53WFVEE285 pKa = 3.83NNPLPGARR293 pKa = 11.84YY294 pKa = 9.24DD295 pKa = 4.45DD296 pKa = 4.67RR297 pKa = 11.84YY298 pKa = 11.29VLQNPDD304 pKa = 4.36EE305 pKa = 4.37IWPPNYY311 pKa = 9.54TDD313 pKa = 4.29ASLSHH318 pKa = 6.74EE319 pKa = 4.36FQLLEE324 pKa = 3.79AWISRR329 pKa = 11.84LSGKK333 pKa = 10.2FSYY336 pKa = 9.43LTGRR340 pKa = 11.84LSYY343 pKa = 10.4SGKK346 pKa = 10.15GSEE349 pKa = 4.16AMSVGTLTDD358 pKa = 3.1SHH360 pKa = 6.87IATTLPQHH368 pKa = 4.93VTISEE373 pKa = 4.32SEE375 pKa = 4.01TNYY378 pKa = 11.3ALDD381 pKa = 4.53IISDD385 pKa = 3.6QSMLLGTLSRR395 pKa = 11.84WRR397 pKa = 11.84EE398 pKa = 3.66YY399 pKa = 10.31PDD401 pKa = 3.77YY402 pKa = 11.33SRR404 pKa = 11.84LPDD407 pKa = 3.35NVTTVDD413 pKa = 3.27PRR415 pKa = 11.84LYY417 pKa = 10.3EE418 pKa = 4.12INSQSGAVRR427 pKa = 11.84KK428 pKa = 9.5IVADD432 pKa = 3.33WHH434 pKa = 6.33ALTAAVLARR443 pKa = 11.84PVV445 pKa = 2.95
MM1 pKa = 7.66DD2 pKa = 4.59TSTSQPSDD10 pKa = 3.44EE11 pKa = 4.79RR12 pKa = 11.84EE13 pKa = 3.92SAPTHH18 pKa = 5.79FRR20 pKa = 11.84SSSNNKK26 pKa = 9.09QNSTNQSGRR35 pKa = 11.84GGARR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84GRR43 pKa = 11.84ASSEE47 pKa = 3.66KK48 pKa = 10.32HH49 pKa = 5.04NEE51 pKa = 3.64AMRR54 pKa = 11.84KK55 pKa = 7.57FHH57 pKa = 7.2GPSQLQAYY65 pKa = 7.93RR66 pKa = 11.84AQLASNNFLDD76 pKa = 5.1LAEE79 pKa = 5.33LSQQLASLASASTPLTVKK97 pKa = 10.76LPATTRR103 pKa = 11.84GLAEE107 pKa = 4.17AVLGGYY113 pKa = 10.25HH114 pKa = 5.5MAFRR118 pKa = 11.84LTKK121 pKa = 10.13EE122 pKa = 4.07PPISLNAFYY131 pKa = 10.58RR132 pKa = 11.84VTLAQFEE139 pKa = 4.39LQAIRR144 pKa = 11.84SQLRR148 pKa = 11.84DD149 pKa = 3.41HH150 pKa = 6.86NISLRR155 pKa = 11.84QYY157 pKa = 9.62HH158 pKa = 5.45YY159 pKa = 11.03EE160 pKa = 3.95EE161 pKa = 5.15FFAKK165 pKa = 10.14YY166 pKa = 10.57ASFEE170 pKa = 4.39QIVAAHH176 pKa = 7.0PATLKK181 pKa = 10.93LVVAVIEE188 pKa = 4.25QYY190 pKa = 9.96GTVSVGGKK198 pKa = 7.82TYY200 pKa = 11.09SPFIVDD206 pKa = 4.9SNVDD210 pKa = 2.87WSVEE214 pKa = 3.52VRR216 pKa = 11.84ARR218 pKa = 11.84RR219 pKa = 11.84RR220 pKa = 11.84GHH222 pKa = 6.4RR223 pKa = 11.84ATGASTSALPHH234 pKa = 6.3RR235 pKa = 11.84VLEE238 pKa = 4.82DD239 pKa = 3.16GGEE242 pKa = 4.27RR243 pKa = 11.84ILEE246 pKa = 4.11VRR248 pKa = 11.84KK249 pKa = 10.62GMGPDD254 pKa = 3.55PYY256 pKa = 10.81TVTFRR261 pKa = 11.84NLEE264 pKa = 3.87NVVRR268 pKa = 11.84VMSDD272 pKa = 3.1EE273 pKa = 4.03NSVLEE278 pKa = 3.94GRR280 pKa = 11.84KK281 pKa = 8.53WFVEE285 pKa = 3.83NNPLPGARR293 pKa = 11.84YY294 pKa = 9.24DD295 pKa = 4.45DD296 pKa = 4.67RR297 pKa = 11.84YY298 pKa = 11.29VLQNPDD304 pKa = 4.36EE305 pKa = 4.37IWPPNYY311 pKa = 9.54TDD313 pKa = 4.29ASLSHH318 pKa = 6.74EE319 pKa = 4.36FQLLEE324 pKa = 3.79AWISRR329 pKa = 11.84LSGKK333 pKa = 10.2FSYY336 pKa = 9.43LTGRR340 pKa = 11.84LSYY343 pKa = 10.4SGKK346 pKa = 10.15GSEE349 pKa = 4.16AMSVGTLTDD358 pKa = 3.1SHH360 pKa = 6.87IATTLPQHH368 pKa = 4.93VTISEE373 pKa = 4.32SEE375 pKa = 4.01TNYY378 pKa = 11.3ALDD381 pKa = 4.53IISDD385 pKa = 3.6QSMLLGTLSRR395 pKa = 11.84WRR397 pKa = 11.84EE398 pKa = 3.66YY399 pKa = 10.31PDD401 pKa = 3.77YY402 pKa = 11.33SRR404 pKa = 11.84LPDD407 pKa = 3.35NVTTVDD413 pKa = 3.27PRR415 pKa = 11.84LYY417 pKa = 10.3EE418 pKa = 4.12INSQSGAVRR427 pKa = 11.84KK428 pKa = 9.5IVADD432 pKa = 3.33WHH434 pKa = 6.33ALTAAVLARR443 pKa = 11.84PVV445 pKa = 2.95
Molecular weight: 49.67 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KLI1|A0A1L3KLI1_9VIRU RdRp OS=Beihai barnacle virus 13 OX=1922357 PE=4 SV=1
MM1 pKa = 7.47FSLPRR6 pKa = 11.84GKK8 pKa = 10.93GFGQYY13 pKa = 9.76HH14 pKa = 5.85AGEE17 pKa = 4.35RR18 pKa = 11.84RR19 pKa = 11.84QTPYY23 pKa = 10.71LPEE26 pKa = 4.0ITDD29 pKa = 3.42IPYY32 pKa = 10.07HH33 pKa = 6.55RR34 pKa = 11.84SAVTLSAIRR43 pKa = 11.84SDD45 pKa = 3.6LKK47 pKa = 11.21QFGTTQKK54 pKa = 10.25IEE56 pKa = 3.94YY57 pKa = 8.94TPTLKK62 pKa = 10.75AAIADD67 pKa = 3.79AYY69 pKa = 10.13KK70 pKa = 10.55EE71 pKa = 3.99FGIGDD76 pKa = 3.76VKK78 pKa = 10.37MLHH81 pKa = 6.63LNDD84 pKa = 2.84VMTYY88 pKa = 10.12KK89 pKa = 10.84GVDD92 pKa = 2.44IWNRR96 pKa = 11.84SPGLPWRR103 pKa = 11.84QHH105 pKa = 4.56GYY107 pKa = 6.99KK108 pKa = 9.74TKK110 pKa = 11.05GEE112 pKa = 4.36VVSDD116 pKa = 3.58MDD118 pKa = 3.75AQRR121 pKa = 11.84SIRR124 pKa = 11.84WFWHH128 pKa = 5.95RR129 pKa = 11.84IKK131 pKa = 10.97DD132 pKa = 3.99GAQMRR137 pKa = 11.84APDD140 pKa = 3.86CSAFTRR146 pKa = 11.84SHH148 pKa = 4.78VCKK151 pKa = 10.62VGEE154 pKa = 4.18EE155 pKa = 4.0KK156 pKa = 10.86VRR158 pKa = 11.84AVWGYY163 pKa = 10.21PMTITMGEE171 pKa = 4.02AVFAIPLIEE180 pKa = 4.44AYY182 pKa = 9.26KK183 pKa = 10.91ARR185 pKa = 11.84GSPIAYY191 pKa = 9.36GYY193 pKa = 9.05EE194 pKa = 3.69ISVGGARR201 pKa = 11.84KK202 pKa = 9.55IFRR205 pKa = 11.84DD206 pKa = 3.49MQTSEE211 pKa = 4.5FKK213 pKa = 10.83AALDD217 pKa = 3.92LEE219 pKa = 4.98SFDD222 pKa = 4.53KK223 pKa = 10.66RR224 pKa = 11.84VPRR227 pKa = 11.84QLIEE231 pKa = 3.78HH232 pKa = 6.93AFDD235 pKa = 4.64ILAQNINFMEE245 pKa = 4.05YY246 pKa = 9.32RR247 pKa = 11.84DD248 pKa = 3.79RR249 pKa = 11.84GIADD253 pKa = 3.07VRR255 pKa = 11.84RR256 pKa = 11.84NIRR259 pKa = 11.84MWEE262 pKa = 3.8YY263 pKa = 10.56LKK265 pKa = 10.67QYY267 pKa = 10.78FLDD270 pKa = 3.93TPVRR274 pKa = 11.84LCTGEE279 pKa = 4.85RR280 pKa = 11.84YY281 pKa = 9.64LKK283 pKa = 10.67RR284 pKa = 11.84GGVPSGSYY292 pKa = 7.81FTQLVDD298 pKa = 4.59SIVNYY303 pKa = 10.24ILVVWLAYY311 pKa = 10.04EE312 pKa = 4.1LTGKK316 pKa = 9.86KK317 pKa = 9.52PRR319 pKa = 11.84YY320 pKa = 8.34LKK322 pKa = 11.17VMGDD326 pKa = 3.68DD327 pKa = 4.55SIAGLDD333 pKa = 3.88SPFHH337 pKa = 6.29LHH339 pKa = 6.36EE340 pKa = 5.25ADD342 pKa = 3.28ALLRR346 pKa = 11.84TVGMKK351 pKa = 10.82LNVPKK356 pKa = 10.66SILSKK361 pKa = 10.88DD362 pKa = 3.68VADD365 pKa = 4.15LTFLGYY371 pKa = 10.41KK372 pKa = 9.12INNGIPSRR380 pKa = 11.84PFDD383 pKa = 3.17KK384 pKa = 10.56WIASLVYY391 pKa = 10.31PEE393 pKa = 5.08TEE395 pKa = 3.99DD396 pKa = 4.77EE397 pKa = 4.31SWDD400 pKa = 3.85DD401 pKa = 3.37VATRR405 pKa = 11.84ALGLLYY411 pKa = 10.54ANCGVDD417 pKa = 3.26SSFDD421 pKa = 3.48RR422 pKa = 11.84LCRR425 pKa = 11.84HH426 pKa = 7.13IINFKK431 pKa = 10.61PFDD434 pKa = 3.66LHH436 pKa = 7.62ISRR439 pKa = 11.84SLSRR443 pKa = 11.84LLWSLGVTGLDD454 pKa = 3.14KK455 pKa = 10.96AVPSSLDD462 pKa = 3.32FYY464 pKa = 11.63LRR466 pKa = 11.84LLL468 pKa = 3.98
MM1 pKa = 7.47FSLPRR6 pKa = 11.84GKK8 pKa = 10.93GFGQYY13 pKa = 9.76HH14 pKa = 5.85AGEE17 pKa = 4.35RR18 pKa = 11.84RR19 pKa = 11.84QTPYY23 pKa = 10.71LPEE26 pKa = 4.0ITDD29 pKa = 3.42IPYY32 pKa = 10.07HH33 pKa = 6.55RR34 pKa = 11.84SAVTLSAIRR43 pKa = 11.84SDD45 pKa = 3.6LKK47 pKa = 11.21QFGTTQKK54 pKa = 10.25IEE56 pKa = 3.94YY57 pKa = 8.94TPTLKK62 pKa = 10.75AAIADD67 pKa = 3.79AYY69 pKa = 10.13KK70 pKa = 10.55EE71 pKa = 3.99FGIGDD76 pKa = 3.76VKK78 pKa = 10.37MLHH81 pKa = 6.63LNDD84 pKa = 2.84VMTYY88 pKa = 10.12KK89 pKa = 10.84GVDD92 pKa = 2.44IWNRR96 pKa = 11.84SPGLPWRR103 pKa = 11.84QHH105 pKa = 4.56GYY107 pKa = 6.99KK108 pKa = 9.74TKK110 pKa = 11.05GEE112 pKa = 4.36VVSDD116 pKa = 3.58MDD118 pKa = 3.75AQRR121 pKa = 11.84SIRR124 pKa = 11.84WFWHH128 pKa = 5.95RR129 pKa = 11.84IKK131 pKa = 10.97DD132 pKa = 3.99GAQMRR137 pKa = 11.84APDD140 pKa = 3.86CSAFTRR146 pKa = 11.84SHH148 pKa = 4.78VCKK151 pKa = 10.62VGEE154 pKa = 4.18EE155 pKa = 4.0KK156 pKa = 10.86VRR158 pKa = 11.84AVWGYY163 pKa = 10.21PMTITMGEE171 pKa = 4.02AVFAIPLIEE180 pKa = 4.44AYY182 pKa = 9.26KK183 pKa = 10.91ARR185 pKa = 11.84GSPIAYY191 pKa = 9.36GYY193 pKa = 9.05EE194 pKa = 3.69ISVGGARR201 pKa = 11.84KK202 pKa = 9.55IFRR205 pKa = 11.84DD206 pKa = 3.49MQTSEE211 pKa = 4.5FKK213 pKa = 10.83AALDD217 pKa = 3.92LEE219 pKa = 4.98SFDD222 pKa = 4.53KK223 pKa = 10.66RR224 pKa = 11.84VPRR227 pKa = 11.84QLIEE231 pKa = 3.78HH232 pKa = 6.93AFDD235 pKa = 4.64ILAQNINFMEE245 pKa = 4.05YY246 pKa = 9.32RR247 pKa = 11.84DD248 pKa = 3.79RR249 pKa = 11.84GIADD253 pKa = 3.07VRR255 pKa = 11.84RR256 pKa = 11.84NIRR259 pKa = 11.84MWEE262 pKa = 3.8YY263 pKa = 10.56LKK265 pKa = 10.67QYY267 pKa = 10.78FLDD270 pKa = 3.93TPVRR274 pKa = 11.84LCTGEE279 pKa = 4.85RR280 pKa = 11.84YY281 pKa = 9.64LKK283 pKa = 10.67RR284 pKa = 11.84GGVPSGSYY292 pKa = 7.81FTQLVDD298 pKa = 4.59SIVNYY303 pKa = 10.24ILVVWLAYY311 pKa = 10.04EE312 pKa = 4.1LTGKK316 pKa = 9.86KK317 pKa = 9.52PRR319 pKa = 11.84YY320 pKa = 8.34LKK322 pKa = 11.17VMGDD326 pKa = 3.68DD327 pKa = 4.55SIAGLDD333 pKa = 3.88SPFHH337 pKa = 6.29LHH339 pKa = 6.36EE340 pKa = 5.25ADD342 pKa = 3.28ALLRR346 pKa = 11.84TVGMKK351 pKa = 10.82LNVPKK356 pKa = 10.66SILSKK361 pKa = 10.88DD362 pKa = 3.68VADD365 pKa = 4.15LTFLGYY371 pKa = 10.41KK372 pKa = 9.12INNGIPSRR380 pKa = 11.84PFDD383 pKa = 3.17KK384 pKa = 10.56WIASLVYY391 pKa = 10.31PEE393 pKa = 5.08TEE395 pKa = 3.99DD396 pKa = 4.77EE397 pKa = 4.31SWDD400 pKa = 3.85DD401 pKa = 3.37VATRR405 pKa = 11.84ALGLLYY411 pKa = 10.54ANCGVDD417 pKa = 3.26SSFDD421 pKa = 3.48RR422 pKa = 11.84LCRR425 pKa = 11.84HH426 pKa = 7.13IINFKK431 pKa = 10.61PFDD434 pKa = 3.66LHH436 pKa = 7.62ISRR439 pKa = 11.84SLSRR443 pKa = 11.84LLWSLGVTGLDD454 pKa = 3.14KK455 pKa = 10.96AVPSSLDD462 pKa = 3.32FYY464 pKa = 11.63LRR466 pKa = 11.84LLL468 pKa = 3.98
Molecular weight: 53.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
913 |
445 |
468 |
456.5 |
51.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.996 ± 0.698 | 0.548 ± 0.385 |
5.805 ± 0.921 | 5.367 ± 0.65 |
3.834 ± 0.483 | 6.572 ± 0.513 |
2.629 ± 0.206 | 4.929 ± 1.095 |
4.491 ± 1.103 | 9.529 ± 0.064 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.081 ± 0.357 | 3.395 ± 0.773 |
4.819 ± 0.088 | 3.395 ± 0.615 |
7.777 ± 0.22 | 8.543 ± 1.419 |
5.586 ± 0.654 | 6.462 ± 0.196 |
1.752 ± 0.284 | 4.491 ± 0.313 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
