
Human smacovirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Huchismacovirus; unclassified Huchismacovirus
Average proteome isoelectric point is 5.77
Get precalculated fractions of proteins
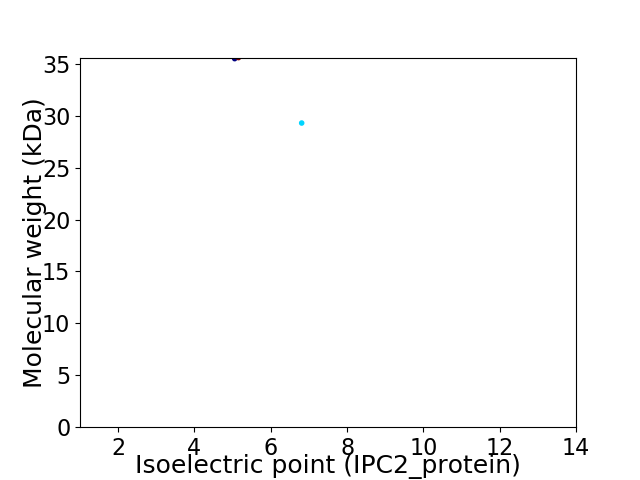
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B5CVG9|A0A0B5CVG9_9VIRU Cap OS=Human smacovirus 1 OX=1595998 GN=Cap PE=4 SV=1
MM1 pKa = 6.74TTQSYY6 pKa = 8.28TYY8 pKa = 10.17FLDD11 pKa = 4.41LATSPDD17 pKa = 3.4KK18 pKa = 11.09VQVVGVSAGGMEE30 pKa = 4.57CMRR33 pKa = 11.84RR34 pKa = 11.84LLPFFAAYY42 pKa = 9.41KK43 pKa = 9.23YY44 pKa = 10.85YY45 pKa = 11.04KK46 pKa = 9.97LGHH49 pKa = 6.08VKK51 pKa = 10.57LKK53 pKa = 10.33VVPASTLPVDD63 pKa = 3.87PTGLSYY69 pKa = 10.68EE70 pKa = 4.23AGEE73 pKa = 4.26NTVDD77 pKa = 5.06PRR79 pKa = 11.84DD80 pKa = 3.46QFNPGLVRR88 pKa = 11.84ITNGEE93 pKa = 4.4DD94 pKa = 2.76IVVPTTMDD102 pKa = 2.85QYY104 pKa = 11.53YY105 pKa = 11.13SLMLDD110 pKa = 3.31RR111 pKa = 11.84RR112 pKa = 11.84WFKK115 pKa = 11.25FNLQSGFSRR124 pKa = 11.84DD125 pKa = 3.09AVPRR129 pKa = 11.84VWRR132 pKa = 11.84IAQLHH137 pKa = 5.29QDD139 pKa = 4.04RR140 pKa = 11.84FPGNTQNFPSVRR152 pKa = 11.84NPLRR156 pKa = 11.84EE157 pKa = 3.87VYY159 pKa = 9.97NRR161 pKa = 11.84QGLVSSADD169 pKa = 3.66PPAVEE174 pKa = 4.26YY175 pKa = 10.36TPLDD179 pKa = 3.73EE180 pKa = 5.93GSDD183 pKa = 3.51PRR185 pKa = 11.84GFFQVGNDD193 pKa = 4.0MYY195 pKa = 11.31LDD197 pKa = 3.92WLPTDD202 pKa = 5.23SDD204 pKa = 4.48TAWDD208 pKa = 3.87TTPNIPKK215 pKa = 9.46YY216 pKa = 9.96GRR218 pKa = 11.84PAAVPEE224 pKa = 4.32VPLMTFILPQAYY236 pKa = 6.99KK237 pKa = 8.99TRR239 pKa = 11.84FYY241 pKa = 10.27YY242 pKa = 9.77RR243 pKa = 11.84VYY245 pKa = 9.5ITEE248 pKa = 3.87TVYY251 pKa = 10.73FKK253 pKa = 11.25EE254 pKa = 4.03PVALNVSSVSSPGSFVVNNGVDD276 pKa = 3.48RR277 pKa = 11.84FIGLPDD283 pKa = 3.24IASGEE288 pKa = 4.11RR289 pKa = 11.84LSSIISASPVYY300 pKa = 10.16SGYY303 pKa = 10.56INDD306 pKa = 3.77GSDD309 pKa = 3.22YY310 pKa = 11.38RR311 pKa = 11.84EE312 pKa = 4.25DD313 pKa = 3.25TGGAA317 pKa = 3.39
MM1 pKa = 6.74TTQSYY6 pKa = 8.28TYY8 pKa = 10.17FLDD11 pKa = 4.41LATSPDD17 pKa = 3.4KK18 pKa = 11.09VQVVGVSAGGMEE30 pKa = 4.57CMRR33 pKa = 11.84RR34 pKa = 11.84LLPFFAAYY42 pKa = 9.41KK43 pKa = 9.23YY44 pKa = 10.85YY45 pKa = 11.04KK46 pKa = 9.97LGHH49 pKa = 6.08VKK51 pKa = 10.57LKK53 pKa = 10.33VVPASTLPVDD63 pKa = 3.87PTGLSYY69 pKa = 10.68EE70 pKa = 4.23AGEE73 pKa = 4.26NTVDD77 pKa = 5.06PRR79 pKa = 11.84DD80 pKa = 3.46QFNPGLVRR88 pKa = 11.84ITNGEE93 pKa = 4.4DD94 pKa = 2.76IVVPTTMDD102 pKa = 2.85QYY104 pKa = 11.53YY105 pKa = 11.13SLMLDD110 pKa = 3.31RR111 pKa = 11.84RR112 pKa = 11.84WFKK115 pKa = 11.25FNLQSGFSRR124 pKa = 11.84DD125 pKa = 3.09AVPRR129 pKa = 11.84VWRR132 pKa = 11.84IAQLHH137 pKa = 5.29QDD139 pKa = 4.04RR140 pKa = 11.84FPGNTQNFPSVRR152 pKa = 11.84NPLRR156 pKa = 11.84EE157 pKa = 3.87VYY159 pKa = 9.97NRR161 pKa = 11.84QGLVSSADD169 pKa = 3.66PPAVEE174 pKa = 4.26YY175 pKa = 10.36TPLDD179 pKa = 3.73EE180 pKa = 5.93GSDD183 pKa = 3.51PRR185 pKa = 11.84GFFQVGNDD193 pKa = 4.0MYY195 pKa = 11.31LDD197 pKa = 3.92WLPTDD202 pKa = 5.23SDD204 pKa = 4.48TAWDD208 pKa = 3.87TTPNIPKK215 pKa = 9.46YY216 pKa = 9.96GRR218 pKa = 11.84PAAVPEE224 pKa = 4.32VPLMTFILPQAYY236 pKa = 6.99KK237 pKa = 8.99TRR239 pKa = 11.84FYY241 pKa = 10.27YY242 pKa = 9.77RR243 pKa = 11.84VYY245 pKa = 9.5ITEE248 pKa = 3.87TVYY251 pKa = 10.73FKK253 pKa = 11.25EE254 pKa = 4.03PVALNVSSVSSPGSFVVNNGVDD276 pKa = 3.48RR277 pKa = 11.84FIGLPDD283 pKa = 3.24IASGEE288 pKa = 4.11RR289 pKa = 11.84LSSIISASPVYY300 pKa = 10.16SGYY303 pKa = 10.56INDD306 pKa = 3.77GSDD309 pKa = 3.22YY310 pKa = 11.38RR311 pKa = 11.84EE312 pKa = 4.25DD313 pKa = 3.25TGGAA317 pKa = 3.39
Molecular weight: 35.49 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5CVG9|A0A0B5CVG9_9VIRU Cap OS=Human smacovirus 1 OX=1595998 GN=Cap PE=4 SV=1
MM1 pKa = 7.68TEE3 pKa = 4.27PKK5 pKa = 9.66WYY7 pKa = 10.02DD8 pKa = 3.0ITVSKK13 pKa = 10.62AKK15 pKa = 10.36CPEE18 pKa = 4.12EE19 pKa = 4.44ILRR22 pKa = 11.84KK23 pKa = 9.22WLDD26 pKa = 3.68EE27 pKa = 4.1NGEE30 pKa = 4.0RR31 pKa = 11.84YY32 pKa = 10.01AYY34 pKa = 10.18GRR36 pKa = 11.84EE37 pKa = 3.94RR38 pKa = 11.84GEE40 pKa = 4.2DD41 pKa = 3.78GYY43 pKa = 11.18EE44 pKa = 3.79HH45 pKa = 6.21FQVRR49 pKa = 11.84VVLRR53 pKa = 11.84NPTSWEE59 pKa = 3.79TMRR62 pKa = 11.84EE63 pKa = 3.68IWGHH67 pKa = 5.79SGHH70 pKa = 6.9CSPTSVRR77 pKa = 11.84NFDD80 pKa = 3.75YY81 pKa = 10.92VLKK84 pKa = 10.38EE85 pKa = 3.68GDD87 pKa = 5.32FICSWIKK94 pKa = 10.79VPDD97 pKa = 4.89AIRR100 pKa = 11.84NAQLRR105 pKa = 11.84PWQQHH110 pKa = 5.67LVDD113 pKa = 5.13LKK115 pKa = 10.9QNDD118 pKa = 3.97RR119 pKa = 11.84EE120 pKa = 4.07VDD122 pKa = 4.03CIIDD126 pKa = 3.68LRR128 pKa = 11.84GNTGKK133 pKa = 10.67SFVTKK138 pKa = 9.01YY139 pKa = 9.83MCMKK143 pKa = 10.56NMAINIPSGLTGKK156 pKa = 10.33DD157 pKa = 3.08IMRR160 pKa = 11.84MCMKK164 pKa = 10.13RR165 pKa = 11.84AKK167 pKa = 10.17LGTYY171 pKa = 9.84IFDD174 pKa = 3.61IPRR177 pKa = 11.84AGSKK181 pKa = 7.45QAKK184 pKa = 5.88EE185 pKa = 3.8TWSAIEE191 pKa = 4.3SIKK194 pKa = 10.51NGYY197 pKa = 7.86MWDD200 pKa = 3.89DD201 pKa = 3.64RR202 pKa = 11.84YY203 pKa = 11.19SWEE206 pKa = 4.16EE207 pKa = 3.53MWIEE211 pKa = 4.07SPRR214 pKa = 11.84VFVFTNEE221 pKa = 3.8YY222 pKa = 9.62PKK224 pKa = 10.97YY225 pKa = 10.4DD226 pKa = 4.21LLSEE230 pKa = 4.02DD231 pKa = 4.74RR232 pKa = 11.84FRR234 pKa = 11.84FWAPGEE240 pKa = 4.14LGLVQLAKK248 pKa = 10.77
MM1 pKa = 7.68TEE3 pKa = 4.27PKK5 pKa = 9.66WYY7 pKa = 10.02DD8 pKa = 3.0ITVSKK13 pKa = 10.62AKK15 pKa = 10.36CPEE18 pKa = 4.12EE19 pKa = 4.44ILRR22 pKa = 11.84KK23 pKa = 9.22WLDD26 pKa = 3.68EE27 pKa = 4.1NGEE30 pKa = 4.0RR31 pKa = 11.84YY32 pKa = 10.01AYY34 pKa = 10.18GRR36 pKa = 11.84EE37 pKa = 3.94RR38 pKa = 11.84GEE40 pKa = 4.2DD41 pKa = 3.78GYY43 pKa = 11.18EE44 pKa = 3.79HH45 pKa = 6.21FQVRR49 pKa = 11.84VVLRR53 pKa = 11.84NPTSWEE59 pKa = 3.79TMRR62 pKa = 11.84EE63 pKa = 3.68IWGHH67 pKa = 5.79SGHH70 pKa = 6.9CSPTSVRR77 pKa = 11.84NFDD80 pKa = 3.75YY81 pKa = 10.92VLKK84 pKa = 10.38EE85 pKa = 3.68GDD87 pKa = 5.32FICSWIKK94 pKa = 10.79VPDD97 pKa = 4.89AIRR100 pKa = 11.84NAQLRR105 pKa = 11.84PWQQHH110 pKa = 5.67LVDD113 pKa = 5.13LKK115 pKa = 10.9QNDD118 pKa = 3.97RR119 pKa = 11.84EE120 pKa = 4.07VDD122 pKa = 4.03CIIDD126 pKa = 3.68LRR128 pKa = 11.84GNTGKK133 pKa = 10.67SFVTKK138 pKa = 9.01YY139 pKa = 9.83MCMKK143 pKa = 10.56NMAINIPSGLTGKK156 pKa = 10.33DD157 pKa = 3.08IMRR160 pKa = 11.84MCMKK164 pKa = 10.13RR165 pKa = 11.84AKK167 pKa = 10.17LGTYY171 pKa = 9.84IFDD174 pKa = 3.61IPRR177 pKa = 11.84AGSKK181 pKa = 7.45QAKK184 pKa = 5.88EE185 pKa = 3.8TWSAIEE191 pKa = 4.3SIKK194 pKa = 10.51NGYY197 pKa = 7.86MWDD200 pKa = 3.89DD201 pKa = 3.64RR202 pKa = 11.84YY203 pKa = 11.19SWEE206 pKa = 4.16EE207 pKa = 3.53MWIEE211 pKa = 4.07SPRR214 pKa = 11.84VFVFTNEE221 pKa = 3.8YY222 pKa = 9.62PKK224 pKa = 10.97YY225 pKa = 10.4DD226 pKa = 4.21LLSEE230 pKa = 4.02DD231 pKa = 4.74RR232 pKa = 11.84FRR234 pKa = 11.84FWAPGEE240 pKa = 4.14LGLVQLAKK248 pKa = 10.77
Molecular weight: 29.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
565 |
248 |
317 |
282.5 |
32.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.133 ± 0.464 | 1.239 ± 0.786 |
6.903 ± 0.3 | 5.664 ± 1.598 |
4.425 ± 0.53 | 6.903 ± 0.3 |
1.062 ± 0.367 | 4.779 ± 1.113 |
4.779 ± 1.65 | 6.726 ± 0.451 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.009 ± 0.681 | 4.248 ± 0.143 |
6.726 ± 1.524 | 3.186 ± 0.242 |
6.903 ± 0.505 | 6.903 ± 0.837 |
5.664 ± 0.817 | 7.611 ± 1.577 |
2.655 ± 1.185 | 5.487 ± 0.7 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
