
Candidatus Aquiluna sp. UB-MaderosW2red
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Luna cluster; Luna-1 subcluster; Aquiluna; unclassified Aquiluna
Average proteome isoelectric point is 6.31
Get precalculated fractions of proteins
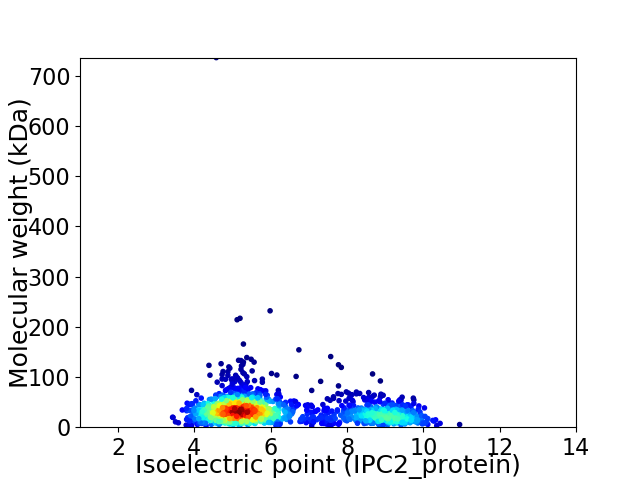
Virtual 2D-PAGE plot for 1613 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G4VLI6|A0A1G4VLI6_9MICO Uncharacterized protein OS=Candidatus Aquiluna sp. UB-MaderosW2red OX=1855377 GN=SAMN05216534_0798 PE=4 SV=1
MM1 pKa = 7.52GKK3 pKa = 9.94YY4 pKa = 9.47LALGAAMLLTLSGCVVSSEE23 pKa = 4.33SGANEE28 pKa = 4.03PEE30 pKa = 4.19DD31 pKa = 3.95AANSAASEE39 pKa = 4.21SSSQQEE45 pKa = 4.15STSQGNGPWAVGEE58 pKa = 4.53LYY60 pKa = 10.51VASDD64 pKa = 3.33HH65 pKa = 5.83TVIINSVRR73 pKa = 11.84TTVEE77 pKa = 3.89SGFSEE82 pKa = 4.31PDD84 pKa = 2.71NDD86 pKa = 5.1FILLIDD92 pKa = 3.65MTIEE96 pKa = 3.77NTGNDD101 pKa = 3.47SHH103 pKa = 7.48SVSSIFGFEE112 pKa = 4.07LQGSDD117 pKa = 4.95DD118 pKa = 3.71YY119 pKa = 11.46MYY121 pKa = 11.04DD122 pKa = 3.03QDD124 pKa = 4.69YY125 pKa = 9.82FADD128 pKa = 3.77TKK130 pKa = 11.19GGLEE134 pKa = 4.12GEE136 pKa = 4.32IAPGSKK142 pKa = 10.28LRR144 pKa = 11.84GEE146 pKa = 4.23IAFDD150 pKa = 3.63VPEE153 pKa = 4.44LEE155 pKa = 4.97FYY157 pKa = 11.22SLLYY161 pKa = 10.83SHH163 pKa = 7.49DD164 pKa = 4.27LFADD168 pKa = 4.25SITFRR173 pKa = 11.84IAASDD178 pKa = 3.9LNIASSAAPEE188 pKa = 4.09EE189 pKa = 4.89SNSSVGLGVGDD200 pKa = 3.78TAEE203 pKa = 4.7GPGFTITLNSVRR215 pKa = 11.84SSEE218 pKa = 4.19EE219 pKa = 3.82DD220 pKa = 3.49SFSGPDD226 pKa = 3.09NDD228 pKa = 5.28FYY230 pKa = 11.59LILDD234 pKa = 3.59MTIEE238 pKa = 4.07NTSDD242 pKa = 3.21EE243 pKa = 4.41EE244 pKa = 4.45ASVSSLASFDD254 pKa = 4.58LKK256 pKa = 11.26GSDD259 pKa = 3.51LYY261 pKa = 10.97QYY263 pKa = 11.11SVGYY267 pKa = 9.09FADD270 pKa = 3.49VKK272 pKa = 11.17GDD274 pKa = 3.77LDD276 pKa = 3.85GSIPAGSKK284 pKa = 10.46LRR286 pKa = 11.84GEE288 pKa = 4.49VAFDD292 pKa = 3.43VPALSFYY299 pKa = 10.88EE300 pKa = 4.12FTYY303 pKa = 10.57KK304 pKa = 10.28HH305 pKa = 6.83DD306 pKa = 4.01FFGSSVTFLIDD317 pKa = 3.94EE318 pKa = 4.65SDD320 pKa = 3.38LGG322 pKa = 3.86
MM1 pKa = 7.52GKK3 pKa = 9.94YY4 pKa = 9.47LALGAAMLLTLSGCVVSSEE23 pKa = 4.33SGANEE28 pKa = 4.03PEE30 pKa = 4.19DD31 pKa = 3.95AANSAASEE39 pKa = 4.21SSSQQEE45 pKa = 4.15STSQGNGPWAVGEE58 pKa = 4.53LYY60 pKa = 10.51VASDD64 pKa = 3.33HH65 pKa = 5.83TVIINSVRR73 pKa = 11.84TTVEE77 pKa = 3.89SGFSEE82 pKa = 4.31PDD84 pKa = 2.71NDD86 pKa = 5.1FILLIDD92 pKa = 3.65MTIEE96 pKa = 3.77NTGNDD101 pKa = 3.47SHH103 pKa = 7.48SVSSIFGFEE112 pKa = 4.07LQGSDD117 pKa = 4.95DD118 pKa = 3.71YY119 pKa = 11.46MYY121 pKa = 11.04DD122 pKa = 3.03QDD124 pKa = 4.69YY125 pKa = 9.82FADD128 pKa = 3.77TKK130 pKa = 11.19GGLEE134 pKa = 4.12GEE136 pKa = 4.32IAPGSKK142 pKa = 10.28LRR144 pKa = 11.84GEE146 pKa = 4.23IAFDD150 pKa = 3.63VPEE153 pKa = 4.44LEE155 pKa = 4.97FYY157 pKa = 11.22SLLYY161 pKa = 10.83SHH163 pKa = 7.49DD164 pKa = 4.27LFADD168 pKa = 4.25SITFRR173 pKa = 11.84IAASDD178 pKa = 3.9LNIASSAAPEE188 pKa = 4.09EE189 pKa = 4.89SNSSVGLGVGDD200 pKa = 3.78TAEE203 pKa = 4.7GPGFTITLNSVRR215 pKa = 11.84SSEE218 pKa = 4.19EE219 pKa = 3.82DD220 pKa = 3.49SFSGPDD226 pKa = 3.09NDD228 pKa = 5.28FYY230 pKa = 11.59LILDD234 pKa = 3.59MTIEE238 pKa = 4.07NTSDD242 pKa = 3.21EE243 pKa = 4.41EE244 pKa = 4.45ASVSSLASFDD254 pKa = 4.58LKK256 pKa = 11.26GSDD259 pKa = 3.51LYY261 pKa = 10.97QYY263 pKa = 11.11SVGYY267 pKa = 9.09FADD270 pKa = 3.49VKK272 pKa = 11.17GDD274 pKa = 3.77LDD276 pKa = 3.85GSIPAGSKK284 pKa = 10.46LRR286 pKa = 11.84GEE288 pKa = 4.49VAFDD292 pKa = 3.43VPALSFYY299 pKa = 10.88EE300 pKa = 4.12FTYY303 pKa = 10.57KK304 pKa = 10.28HH305 pKa = 6.83DD306 pKa = 4.01FFGSSVTFLIDD317 pKa = 3.94EE318 pKa = 4.65SDD320 pKa = 3.38LGG322 pKa = 3.86
Molecular weight: 34.29 kDa
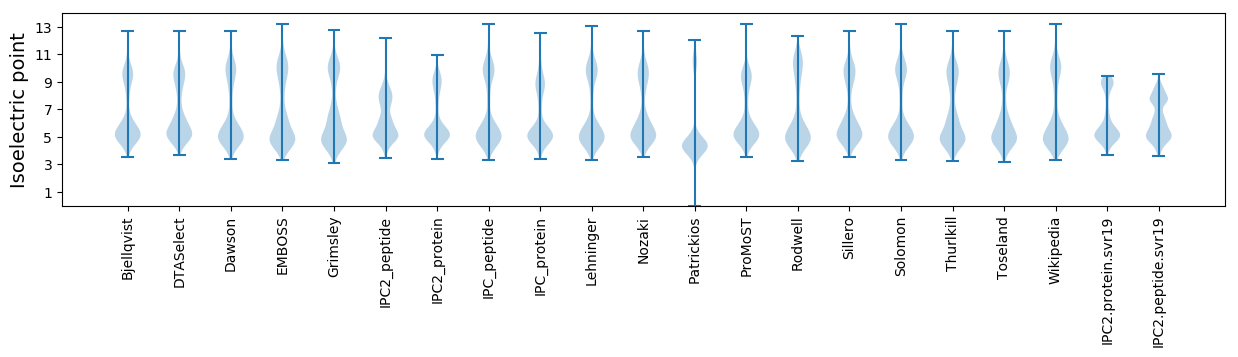
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G4VC75|A0A1G4VC75_9MICO Chromosome partitioning protein OS=Candidatus Aquiluna sp. UB-MaderosW2red OX=1855377 GN=SAMN05216534_0263 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 10.04KK16 pKa = 8.86HH17 pKa = 4.34GFRR20 pKa = 11.84ARR22 pKa = 11.84MHH24 pKa = 5.46TRR26 pKa = 11.84AGRR29 pKa = 11.84AILAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.65GRR40 pKa = 11.84SEE42 pKa = 3.94LSAA45 pKa = 4.73
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 10.04KK16 pKa = 8.86HH17 pKa = 4.34GFRR20 pKa = 11.84ARR22 pKa = 11.84MHH24 pKa = 5.46TRR26 pKa = 11.84AGRR29 pKa = 11.84AILAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.65GRR40 pKa = 11.84SEE42 pKa = 3.94LSAA45 pKa = 4.73
Molecular weight: 5.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
512337 |
25 |
7338 |
317.6 |
34.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.561 ± 0.067 | 0.536 ± 0.013 |
5.111 ± 0.043 | 6.218 ± 0.086 |
3.951 ± 0.042 | 8.219 ± 0.085 |
1.674 ± 0.031 | 6.245 ± 0.045 |
4.412 ± 0.057 | 10.804 ± 0.104 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.149 ± 0.035 | 3.299 ± 0.05 |
4.414 ± 0.033 | 3.569 ± 0.034 |
5.211 ± 0.065 | 7.036 ± 0.084 |
5.453 ± 0.123 | 7.686 ± 0.057 |
1.242 ± 0.03 | 2.21 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
