
Pacific flying fox faeces associated circular DNA virus-15
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.16
Get precalculated fractions of proteins
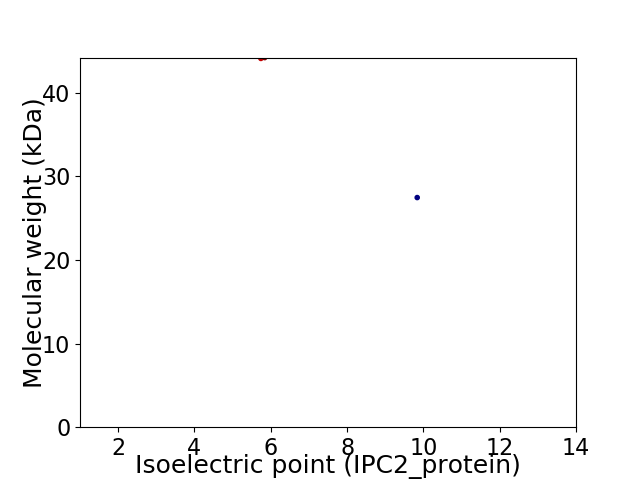
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140CTW4|A0A140CTW4_9VIRU Putative capsid protein OS=Pacific flying fox faeces associated circular DNA virus-15 OX=1796010 PE=4 SV=1
MM1 pKa = 7.69DD2 pKa = 4.21TFLTAHH8 pKa = 5.72QEE10 pKa = 4.0EE11 pKa = 5.04EE12 pKa = 3.88PRR14 pKa = 11.84YY15 pKa = 9.01PGSDD19 pKa = 3.5VEE21 pKa = 5.8DD22 pKa = 5.14DD23 pKa = 4.55SLLCLEE29 pKa = 5.44GPLSPEE35 pKa = 4.47DD36 pKa = 3.74EE37 pKa = 4.23TLEE40 pKa = 4.28APKK43 pKa = 10.27EE44 pKa = 4.15KK45 pKa = 10.29PKK47 pKa = 10.79SEE49 pKa = 4.01KK50 pKa = 10.3KK51 pKa = 10.16KK52 pKa = 10.37GSFRR56 pKa = 11.84LRR58 pKa = 11.84GTQFILTFPQCDD70 pKa = 3.43VKK72 pKa = 11.28KK73 pKa = 10.06EE74 pKa = 3.97VAAEE78 pKa = 4.36RR79 pKa = 11.84IDD81 pKa = 3.81IAWGKK86 pKa = 10.31DD87 pKa = 2.83LTGYY91 pKa = 7.57VVCEE95 pKa = 4.14EE96 pKa = 3.98NHH98 pKa = 6.93KK99 pKa = 11.14DD100 pKa = 3.76GTPHH104 pKa = 6.0LHH106 pKa = 6.47VYY108 pKa = 10.74LKK110 pKa = 10.76FEE112 pKa = 4.08NRR114 pKa = 11.84KK115 pKa = 9.5SFSKK119 pKa = 10.63VDD121 pKa = 3.46CFDD124 pKa = 5.64FIGQKK129 pKa = 9.69HH130 pKa = 5.79GNYY133 pKa = 9.29QVVRR137 pKa = 11.84STLKK141 pKa = 10.3SIEE144 pKa = 4.12YY145 pKa = 7.57VTKK148 pKa = 10.49GGNYY152 pKa = 7.25VAKK155 pKa = 10.24GVDD158 pKa = 3.27VEE160 pKa = 4.64SIKK163 pKa = 10.75KK164 pKa = 10.01KK165 pKa = 10.18KK166 pKa = 9.97APKK169 pKa = 8.5NQEE172 pKa = 3.64IAKK175 pKa = 8.89MLLDD179 pKa = 4.32GKK181 pKa = 11.36SMAEE185 pKa = 3.89INEE188 pKa = 4.17DD189 pKa = 3.04HH190 pKa = 6.97CGYY193 pKa = 11.67VMINKK198 pKa = 9.75RR199 pKa = 11.84KK200 pKa = 8.76IEE202 pKa = 4.18EE203 pKa = 4.02YY204 pKa = 8.22EE205 pKa = 4.01TWVKK209 pKa = 10.63CEE211 pKa = 4.36KK212 pKa = 10.13IKK214 pKa = 10.62KK215 pKa = 10.14SKK217 pKa = 10.18IDD219 pKa = 4.06WIPPSLDD226 pKa = 2.94GLTDD230 pKa = 3.83ANLQIATWICSNIRR244 pKa = 11.84QNRR247 pKa = 11.84AFKK250 pKa = 10.67APQLYY255 pKa = 9.78IHH257 pKa = 7.06GPRR260 pKa = 11.84NLGKK264 pKa = 8.97TSLVEE269 pKa = 3.61WLEE272 pKa = 4.14RR273 pKa = 11.84SLSVYY278 pKa = 10.53HH279 pKa = 6.21MPTMEE284 pKa = 5.29DD285 pKa = 4.19FYY287 pKa = 11.65DD288 pKa = 4.75LYY290 pKa = 11.43SDD292 pKa = 5.57DD293 pKa = 4.25YY294 pKa = 11.07DD295 pKa = 3.96LVVIDD300 pKa = 4.45EE301 pKa = 4.76FKK303 pKa = 10.78GQKK306 pKa = 8.92TIQFLNQFLQGSVMPIRR323 pKa = 11.84KK324 pKa = 9.42KK325 pKa = 10.76GSQGDD330 pKa = 3.5KK331 pKa = 10.7SKK333 pKa = 10.95NLPVVILSNYY343 pKa = 7.45TLSEE347 pKa = 4.64CYY349 pKa = 10.42VKK351 pKa = 10.68AANDD355 pKa = 3.31GRR357 pKa = 11.84LNTLEE362 pKa = 4.07CRR364 pKa = 11.84LEE366 pKa = 3.97IVEE369 pKa = 4.24VDD371 pKa = 3.83SFIDD375 pKa = 4.29FYY377 pKa = 11.55KK378 pKa = 10.84DD379 pKa = 3.16RR380 pKa = 11.84NDD382 pKa = 4.63LII384 pKa = 5.54
MM1 pKa = 7.69DD2 pKa = 4.21TFLTAHH8 pKa = 5.72QEE10 pKa = 4.0EE11 pKa = 5.04EE12 pKa = 3.88PRR14 pKa = 11.84YY15 pKa = 9.01PGSDD19 pKa = 3.5VEE21 pKa = 5.8DD22 pKa = 5.14DD23 pKa = 4.55SLLCLEE29 pKa = 5.44GPLSPEE35 pKa = 4.47DD36 pKa = 3.74EE37 pKa = 4.23TLEE40 pKa = 4.28APKK43 pKa = 10.27EE44 pKa = 4.15KK45 pKa = 10.29PKK47 pKa = 10.79SEE49 pKa = 4.01KK50 pKa = 10.3KK51 pKa = 10.16KK52 pKa = 10.37GSFRR56 pKa = 11.84LRR58 pKa = 11.84GTQFILTFPQCDD70 pKa = 3.43VKK72 pKa = 11.28KK73 pKa = 10.06EE74 pKa = 3.97VAAEE78 pKa = 4.36RR79 pKa = 11.84IDD81 pKa = 3.81IAWGKK86 pKa = 10.31DD87 pKa = 2.83LTGYY91 pKa = 7.57VVCEE95 pKa = 4.14EE96 pKa = 3.98NHH98 pKa = 6.93KK99 pKa = 11.14DD100 pKa = 3.76GTPHH104 pKa = 6.0LHH106 pKa = 6.47VYY108 pKa = 10.74LKK110 pKa = 10.76FEE112 pKa = 4.08NRR114 pKa = 11.84KK115 pKa = 9.5SFSKK119 pKa = 10.63VDD121 pKa = 3.46CFDD124 pKa = 5.64FIGQKK129 pKa = 9.69HH130 pKa = 5.79GNYY133 pKa = 9.29QVVRR137 pKa = 11.84STLKK141 pKa = 10.3SIEE144 pKa = 4.12YY145 pKa = 7.57VTKK148 pKa = 10.49GGNYY152 pKa = 7.25VAKK155 pKa = 10.24GVDD158 pKa = 3.27VEE160 pKa = 4.64SIKK163 pKa = 10.75KK164 pKa = 10.01KK165 pKa = 10.18KK166 pKa = 9.97APKK169 pKa = 8.5NQEE172 pKa = 3.64IAKK175 pKa = 8.89MLLDD179 pKa = 4.32GKK181 pKa = 11.36SMAEE185 pKa = 3.89INEE188 pKa = 4.17DD189 pKa = 3.04HH190 pKa = 6.97CGYY193 pKa = 11.67VMINKK198 pKa = 9.75RR199 pKa = 11.84KK200 pKa = 8.76IEE202 pKa = 4.18EE203 pKa = 4.02YY204 pKa = 8.22EE205 pKa = 4.01TWVKK209 pKa = 10.63CEE211 pKa = 4.36KK212 pKa = 10.13IKK214 pKa = 10.62KK215 pKa = 10.14SKK217 pKa = 10.18IDD219 pKa = 4.06WIPPSLDD226 pKa = 2.94GLTDD230 pKa = 3.83ANLQIATWICSNIRR244 pKa = 11.84QNRR247 pKa = 11.84AFKK250 pKa = 10.67APQLYY255 pKa = 9.78IHH257 pKa = 7.06GPRR260 pKa = 11.84NLGKK264 pKa = 8.97TSLVEE269 pKa = 3.61WLEE272 pKa = 4.14RR273 pKa = 11.84SLSVYY278 pKa = 10.53HH279 pKa = 6.21MPTMEE284 pKa = 5.29DD285 pKa = 4.19FYY287 pKa = 11.65DD288 pKa = 4.75LYY290 pKa = 11.43SDD292 pKa = 5.57DD293 pKa = 4.25YY294 pKa = 11.07DD295 pKa = 3.96LVVIDD300 pKa = 4.45EE301 pKa = 4.76FKK303 pKa = 10.78GQKK306 pKa = 8.92TIQFLNQFLQGSVMPIRR323 pKa = 11.84KK324 pKa = 9.42KK325 pKa = 10.76GSQGDD330 pKa = 3.5KK331 pKa = 10.7SKK333 pKa = 10.95NLPVVILSNYY343 pKa = 7.45TLSEE347 pKa = 4.64CYY349 pKa = 10.42VKK351 pKa = 10.68AANDD355 pKa = 3.31GRR357 pKa = 11.84LNTLEE362 pKa = 4.07CRR364 pKa = 11.84LEE366 pKa = 3.97IVEE369 pKa = 4.24VDD371 pKa = 3.83SFIDD375 pKa = 4.29FYY377 pKa = 11.55KK378 pKa = 10.84DD379 pKa = 3.16RR380 pKa = 11.84NDD382 pKa = 4.63LII384 pKa = 5.54
Molecular weight: 44.12 kDa
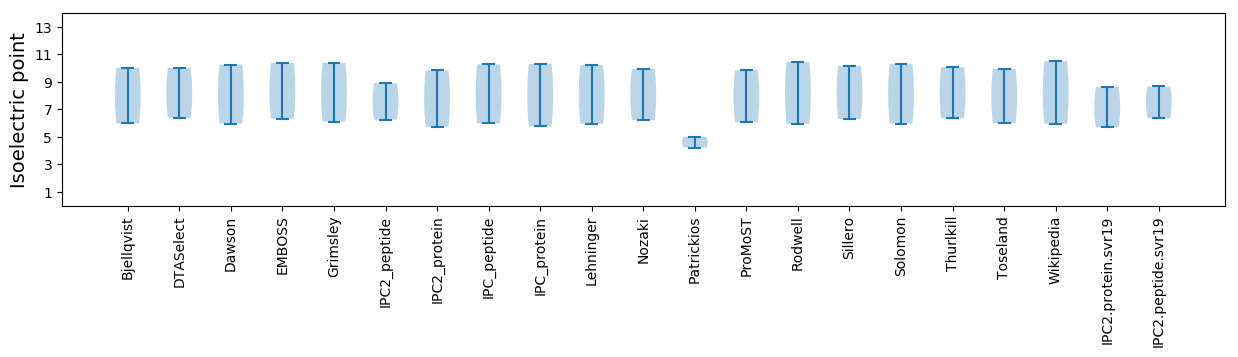
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140CTW4|A0A140CTW4_9VIRU Putative capsid protein OS=Pacific flying fox faeces associated circular DNA virus-15 OX=1796010 PE=4 SV=1
MM1 pKa = 7.01VGYY4 pKa = 10.46VRR6 pKa = 11.84TITYY10 pKa = 8.53RR11 pKa = 11.84RR12 pKa = 11.84PYY14 pKa = 9.26GSVYY18 pKa = 10.66GPYY21 pKa = 10.41NKK23 pKa = 9.83YY24 pKa = 10.08ARR26 pKa = 11.84SRR28 pKa = 11.84RR29 pKa = 11.84IVNTAAKK36 pKa = 10.18LLRR39 pKa = 11.84KK40 pKa = 9.96RR41 pKa = 11.84MGGGPRR47 pKa = 11.84APLATRR53 pKa = 11.84GWYY56 pKa = 10.31GNTYY60 pKa = 9.4GKK62 pKa = 10.46RR63 pKa = 11.84GGEE66 pKa = 3.84LKK68 pKa = 11.01YY69 pKa = 10.33IDD71 pKa = 4.43VDD73 pKa = 3.82NVPAALPVGGTVDD86 pKa = 3.62VLNAIASGAGIDD98 pKa = 3.77EE99 pKa = 4.15RR100 pKa = 11.84VGRR103 pKa = 11.84QITLKK108 pKa = 10.71SIFSRR113 pKa = 11.84WGFYY117 pKa = 10.49PSTTTSAPQGTIVRR131 pKa = 11.84VLIVYY136 pKa = 8.02DD137 pKa = 3.74AQANSAASPPAVNEE151 pKa = 3.8ILATGTWDD159 pKa = 4.5SPMNLNNRR167 pKa = 11.84EE168 pKa = 3.83RR169 pKa = 11.84FKK171 pKa = 10.77IITEE175 pKa = 4.19FKK177 pKa = 8.16VTVGATLYY185 pKa = 9.92TAGALTAGSPVPRR198 pKa = 11.84VVEE201 pKa = 4.4KK202 pKa = 10.94YY203 pKa = 10.22KK204 pKa = 10.57KK205 pKa = 10.69LNMTTTYY212 pKa = 10.66SGPNNTNANIATGAIYY228 pKa = 10.68VVTIPSLNNTVVADD242 pKa = 3.81YY243 pKa = 9.3YY244 pKa = 11.21HH245 pKa = 6.69RR246 pKa = 11.84TRR248 pKa = 11.84YY249 pKa = 8.48MDD251 pKa = 3.49CC252 pKa = 4.13
MM1 pKa = 7.01VGYY4 pKa = 10.46VRR6 pKa = 11.84TITYY10 pKa = 8.53RR11 pKa = 11.84RR12 pKa = 11.84PYY14 pKa = 9.26GSVYY18 pKa = 10.66GPYY21 pKa = 10.41NKK23 pKa = 9.83YY24 pKa = 10.08ARR26 pKa = 11.84SRR28 pKa = 11.84RR29 pKa = 11.84IVNTAAKK36 pKa = 10.18LLRR39 pKa = 11.84KK40 pKa = 9.96RR41 pKa = 11.84MGGGPRR47 pKa = 11.84APLATRR53 pKa = 11.84GWYY56 pKa = 10.31GNTYY60 pKa = 9.4GKK62 pKa = 10.46RR63 pKa = 11.84GGEE66 pKa = 3.84LKK68 pKa = 11.01YY69 pKa = 10.33IDD71 pKa = 4.43VDD73 pKa = 3.82NVPAALPVGGTVDD86 pKa = 3.62VLNAIASGAGIDD98 pKa = 3.77EE99 pKa = 4.15RR100 pKa = 11.84VGRR103 pKa = 11.84QITLKK108 pKa = 10.71SIFSRR113 pKa = 11.84WGFYY117 pKa = 10.49PSTTTSAPQGTIVRR131 pKa = 11.84VLIVYY136 pKa = 8.02DD137 pKa = 3.74AQANSAASPPAVNEE151 pKa = 3.8ILATGTWDD159 pKa = 4.5SPMNLNNRR167 pKa = 11.84EE168 pKa = 3.83RR169 pKa = 11.84FKK171 pKa = 10.77IITEE175 pKa = 4.19FKK177 pKa = 8.16VTVGATLYY185 pKa = 9.92TAGALTAGSPVPRR198 pKa = 11.84VVEE201 pKa = 4.4KK202 pKa = 10.94YY203 pKa = 10.22KK204 pKa = 10.57KK205 pKa = 10.69LNMTTTYY212 pKa = 10.66SGPNNTNANIATGAIYY228 pKa = 10.68VVTIPSLNNTVVADD242 pKa = 3.81YY243 pKa = 9.3YY244 pKa = 11.21HH245 pKa = 6.69RR246 pKa = 11.84TRR248 pKa = 11.84YY249 pKa = 8.48MDD251 pKa = 3.49CC252 pKa = 4.13
Molecular weight: 27.48 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
636 |
252 |
384 |
318.0 |
35.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.289 ± 2.21 | 1.572 ± 0.716 |
5.818 ± 1.609 | 5.975 ± 2.188 |
2.987 ± 0.852 | 7.233 ± 1.395 |
1.415 ± 0.62 | 6.132 ± 0.109 |
8.176 ± 2.32 | 7.39 ± 1.117 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.887 ± 0.059 | 5.346 ± 0.852 |
4.874 ± 0.656 | 2.673 ± 0.902 |
5.503 ± 1.481 | 5.818 ± 0.401 |
6.761 ± 2.165 | 7.547 ± 0.962 |
1.258 ± 0.041 | 5.346 ± 1.094 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
