
Mycobacterium pseudokansasii
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Corynebacteriales; Mycobacteriaceae; Mycobacterium
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
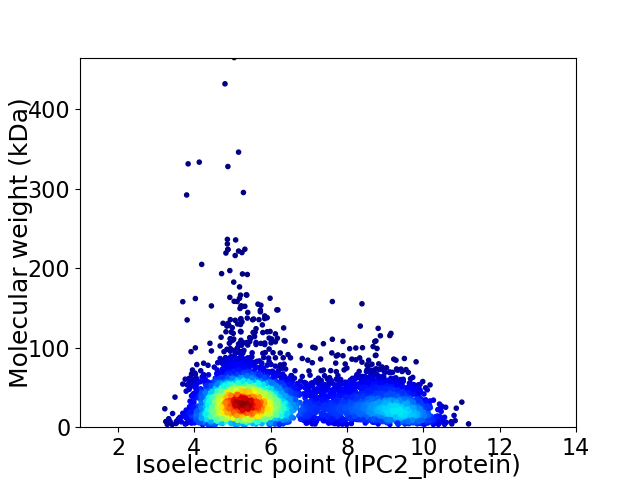
Virtual 2D-PAGE plot for 5730 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A498QZP5|A0A498QZP5_9MYCO Non-specific serine/threonine protein kinase OS=Mycobacterium pseudokansasii OX=2341080 GN=pknJ_2 PE=4 SV=1
MM1 pKa = 7.23NFSVSPPEE9 pKa = 3.89INSARR14 pKa = 11.84IFSGAGAGPMLAAAAAWDD32 pKa = 3.77QLAGEE37 pKa = 4.95LGSAATAFSSVTSALVSSSWQGPASAAMASVAGGYY72 pKa = 10.01LGWLSSAGAQAAQAGGQARR91 pKa = 11.84IAAAAFEE98 pKa = 4.41ATRR101 pKa = 11.84AATVHH106 pKa = 6.49PAAVWANRR114 pKa = 11.84SQLVSLVASDD124 pKa = 4.62LLGFNAPAIAAVEE137 pKa = 3.85AAYY140 pKa = 10.15EE141 pKa = 4.06QMWAQDD147 pKa = 3.41VAAMFGYY154 pKa = 10.28HH155 pKa = 6.69AGASAAASALTPFIQLVQNPVASGEE180 pKa = 4.19ALVAAAQAAILSPPGRR196 pKa = 11.84VSIFNAGLADD206 pKa = 3.58VGVGNVGFGSVGDD219 pKa = 4.08GNVGAGNVGGSNVGFGNLGSLNFGSGNLGSFNVGSGNIGLYY260 pKa = 10.71NFGPGNLGSYY270 pKa = 10.35NVGLGNAGDD279 pKa = 3.91YY280 pKa = 11.39NFGFGNTGIGNIGFGNTGSHH300 pKa = 6.86NIGIGLTGDD309 pKa = 3.63GQVGFGGWNSGSANVGLFNSGIGNVGLFNSGTGNWGVGNSGEE351 pKa = 4.44LNTGLFNPGRR361 pKa = 11.84LNTGVFNTGLLNTGVGNAGSYY382 pKa = 10.96NSGSFNVGASNTGSWNAGDD401 pKa = 4.44TNTGWFNPGDD411 pKa = 3.9VNTGIGNTGDD421 pKa = 3.52VNTGGFNQGNLNNGFFWRR439 pKa = 11.84GDD441 pKa = 3.83GQGHH445 pKa = 7.23AGFDD449 pKa = 3.62YY450 pKa = 10.74TLTIPAIGLNLGADD464 pKa = 3.89VPLGIPVTGTIGTIVNGGPPEE485 pKa = 3.79ITVPGFTIPTLHH497 pKa = 6.29LTGDD501 pKa = 3.87ALFGTIGPIVVDD513 pKa = 5.16PITITGPSLNLTVGGGQSLQLGFSGPGVGPVVIPVLQVAAGPGVGNSTGVPSSGFFNSGVGGASGFGNVGGGSGWWNFGRR593 pKa = 11.84SSGVGNVGVLGSGVLNLGDD612 pKa = 4.49GVSGWFNTIPSMGSGVGNVGEE633 pKa = 4.03QLAGLFSAGPGGPSVFNLGLGNQGGLNLGHH663 pKa = 7.08ANVGDD668 pKa = 3.95FNLGGGNVGDD678 pKa = 4.6HH679 pKa = 6.15NVGGANVGDD688 pKa = 4.09ANVGVGNIGDD698 pKa = 4.11HH699 pKa = 5.98NVGGGNVGNVNVGGGNAGDD718 pKa = 4.36GNRR721 pKa = 11.84GWGNTGSFNVGFGNTGVGNFGLANQGSNNIGIGLSGDD758 pKa = 3.47NQIGFGGFNSGAGNVGLFNSGSNNIGFFNSGTGNFGVGNSGSFNTGIASTGSTNTGVFNTGLGNTGWANAGDD830 pKa = 4.11FNSGGFNAGAFNTGSFNPGDD850 pKa = 3.95TNTGWFNIGDD860 pKa = 4.44LNTGAFNTGDD870 pKa = 3.43INTGAFISGDD880 pKa = 3.49ANNGFFWRR888 pKa = 11.84GTGQGLIAADD898 pKa = 3.65YY899 pKa = 9.11TLTIPHH905 pKa = 6.92IPLTLGGGGFFEE917 pKa = 5.26LPITGSITGMTVQSFMVHH935 pKa = 7.04GIGDD939 pKa = 4.46SNTGIPLNLQLNVLGQQTDD958 pKa = 3.77EE959 pKa = 4.88IGVQVHH965 pKa = 6.14IPVIDD970 pKa = 3.8KK971 pKa = 10.8DD972 pKa = 3.87VTIPLPALPISLGITVDD989 pKa = 3.63NQIGAILVPPITINPITFNNVMVGNNTTSLSADD1022 pKa = 3.32VAGGIGPVTIPVFQLAAAPGFGNITGVPSSGFFNSGVGGGSGFGNSGSGVSGWWNVASLSSGYY1085 pKa = 10.38QNLGNLVSGFINKK1098 pKa = 9.69GEE1100 pKa = 4.1LLSGINNSNMLGLSTSGVGNVGDD1123 pKa = 3.93NVSGLFFQGADD1134 pKa = 2.57RR1135 pKa = 11.84VSIFNAGLADD1145 pKa = 3.58VGVGNVGFGSVGDD1158 pKa = 4.08GNVGAGNVGGSNVGFGNLGSLNFGSGNLGSFNVGSGNIGLYY1199 pKa = 10.71NFGPGNLGSYY1209 pKa = 10.35NVGLGNAGDD1218 pKa = 3.91YY1219 pKa = 11.39NFGFGNTGIGNIGFGNTGSHH1239 pKa = 6.86NIGIGLTGDD1248 pKa = 3.63GQVGFGGWNSGSANVGLFNSGAGNVGLFNSGTGNWGVGNSGEE1290 pKa = 4.44LNTGLFNPGRR1300 pKa = 11.84LNTGVFNTGLLNTGVGNAGSYY1321 pKa = 10.96NSGSFNVGASNTGSWNAGDD1340 pKa = 4.44TNTGWFNPGDD1350 pKa = 3.9VNTGIGNTGDD1360 pKa = 3.52VNTGGFNQGNLNNGFFWRR1378 pKa = 11.84GDD1380 pKa = 3.83GQGHH1384 pKa = 7.23AGFDD1388 pKa = 3.62YY1389 pKa = 10.74TLTIPAIALNLGVNVPLNIPITGHH1413 pKa = 6.75LGDD1416 pKa = 4.03IVIGSFTIPTLHH1428 pKa = 6.93LNGSNLTGTIGPIVVDD1444 pKa = 5.16PITITGPSLDD1454 pKa = 3.99LHH1456 pKa = 6.88VGGPGEE1462 pKa = 4.25SLQLDD1467 pKa = 3.72ISGPALGPVVIPVLQVAAGPGVGNSTGVPSSGFFNSGVGGASGFGNVGGGSGWWNFGGYY1526 pKa = 9.71SGVGNVGVLGSGVLNLGDD1544 pKa = 4.49GVSGWFNTVPSMGSGVGNVGEE1565 pKa = 4.03QLAGLFSAGPGGPSVFNLGLGNQGGLNLGHH1595 pKa = 7.08ANVGDD1600 pKa = 3.95FNLGGGNVGDD1610 pKa = 4.6HH1611 pKa = 6.15NVGGANVGDD1620 pKa = 4.09ANVGVGNIGDD1630 pKa = 4.11HH1631 pKa = 5.98NVGGGNVGNVNVGGSNAGDD1650 pKa = 3.88GNRR1653 pKa = 11.84GWGNTGSFNVGFGNTGVGNFGLANQGSNNIGIGLSGDD1690 pKa = 3.47NQIGFGGFNSGAGNVGLFNSGSNNIGFFNSGTGNFGVGNSGSFNTGIASTGSTNTGVFNTGLGNTGWANAGDD1762 pKa = 4.11FNSGGFNAGAFNTGSFNPGDD1782 pKa = 3.95TNTGWFNIGDD1792 pKa = 4.44LNTGAFNTGDD1802 pKa = 3.43INTGAFISGDD1812 pKa = 3.49ANNGFFWRR1820 pKa = 11.84GTGQGLIAADD1830 pKa = 3.65YY1831 pKa = 9.11TLTIPHH1837 pKa = 6.88IPLTLGLGGKK1847 pKa = 9.76LDD1849 pKa = 3.75IPITGQIAGLTVNPITLHH1867 pKa = 6.33GIGGSTTGIPVNAHH1881 pKa = 4.82FTLLGSTGGTTIHH1894 pKa = 6.38FNIPGTDD1901 pKa = 3.12LGFSFDD1907 pKa = 4.7LPSLPISIGVDD1918 pKa = 2.43IGDD1921 pKa = 3.76QIGAITIPQITINPITFNNVVVGGDD1946 pKa = 3.72TTSLSADD1953 pKa = 3.35VAGGIGPVTIPVFQLAAAPGFGNITGVPSSGFFNSGVGGGSGFGNSGSGVSGWWNVASLSSGYY2016 pKa = 10.38QNLGNLVSGFINKK2029 pKa = 9.69GEE2031 pKa = 4.1LLSGINNSNMLGLSTSGVGNVGDD2054 pKa = 3.93NVSGLFFQGADD2065 pKa = 2.57RR2066 pKa = 11.84VSIFNAGLADD2076 pKa = 3.58VGVGNVGFGSVGDD2089 pKa = 4.08GNVGAGNVGGSNVGFGNLGSLNFGSGNLGSFNVGSGNIGLYY2130 pKa = 10.71NFGPGNLGSYY2140 pKa = 10.35NVGLGNAGDD2149 pKa = 3.91YY2150 pKa = 11.39NFGFGNTGIGNIGFGNTGSHH2170 pKa = 6.86NIGIGLTGDD2179 pKa = 3.63GQVGFGGWNSGSANVGLFNSGAGNVGLFNSGTGNWGVGNSGEE2221 pKa = 4.44LNTGLFNPGRR2231 pKa = 11.84LNTGVFNTGLLNTGVGNAGSYY2252 pKa = 10.96NSGSFNVGASNTGSWNAGDD2271 pKa = 4.44TNTGWFNPGDD2281 pKa = 3.9VNTGIGNTGDD2291 pKa = 3.52VNTGGFNQGNLNNGFFWRR2309 pKa = 11.84GDD2311 pKa = 3.83GQGHH2315 pKa = 7.23AGFDD2319 pKa = 3.62YY2320 pKa = 10.74TLTIPAIALNLGVNVPLNIPITGHH2344 pKa = 6.63LGDD2347 pKa = 4.07IVVDD2351 pKa = 3.99PFTIPVLHH2359 pKa = 6.22LTGTGGNSLTGTIGPIVSDD2378 pKa = 3.89QITITGPSLNLTLGGPGEE2396 pKa = 4.13SLQLGFSGPALGPVVIPVLQVAAGPGVGNSTGVPSSGFFNSGVGGASGFGNVGGGSGWWNFGGYY2460 pKa = 9.71SGVGNVGVLGSGVLNLGDD2478 pKa = 4.49GVSGWFNTVPSMGSGVGNVGEE2499 pKa = 4.03QLAGLFSAGPGGPSVFNLGLGNQGGLNLGHH2529 pKa = 7.08ANVGDD2534 pKa = 3.95FNLGGGNVGDD2544 pKa = 4.6HH2545 pKa = 6.15NVGGANVGDD2554 pKa = 4.09ANVGVGNIGDD2564 pKa = 4.11HH2565 pKa = 5.98NVGGGNVGNVNVGGGNAGDD2584 pKa = 4.36GNRR2587 pKa = 11.84GWGNTGSFNVGFGNTGVGNFGLANQGSNNIGIGLSGDD2624 pKa = 3.47NQIGFGGFNSGAGNVGLFNSGSNNIGFFNSGTGNFGVGNSGSFNTGIASTGSTNTGVFNTGLGNTGWANAGDD2696 pKa = 4.11FNSGGFNAGAFNTGSFNPGDD2716 pKa = 3.95TNTGWFNIGDD2726 pKa = 4.44LNTGAFNTGDD2736 pKa = 3.43INTGAFISGDD2746 pKa = 3.49ANNGFFWRR2754 pKa = 11.84GTGQGLIAADD2764 pKa = 3.65YY2765 pKa = 9.11TLTIPHH2771 pKa = 6.88IPLTLGLGGKK2781 pKa = 9.76LDD2783 pKa = 3.75IPITGQIAGLTVNQFTLHH2801 pKa = 6.46GEE2803 pKa = 4.08NGPGIPLNLLLAIDD2817 pKa = 4.57ADD2819 pKa = 4.06SGGTSIHH2826 pKa = 6.38VGIPDD2831 pKa = 3.55TDD2833 pKa = 3.17IGFDD2837 pKa = 3.47IPIDD2841 pKa = 3.91GLPITLTLPLNSEE2854 pKa = 4.53LSPIVIEE2861 pKa = 5.08PITIGAIPLDD2871 pKa = 3.94LTVGGDD2877 pKa = 3.45TTQLGTALAAGIGPITATLFHH2898 pKa = 6.78VSPVPGFGNSTGSPSSGFFNSGAGGSSGFGNIGDD2932 pKa = 4.1TLSGFWNVGSEE2943 pKa = 4.18GSGFEE2948 pKa = 4.09NYY2950 pKa = 10.03GGSLLSGITNLGGSLSGIDD2969 pKa = 3.51NTSSLGLALAGVVSGLGNIGSQLSGLFLSGSVPP3002 pKa = 3.3
MM1 pKa = 7.23NFSVSPPEE9 pKa = 3.89INSARR14 pKa = 11.84IFSGAGAGPMLAAAAAWDD32 pKa = 3.77QLAGEE37 pKa = 4.95LGSAATAFSSVTSALVSSSWQGPASAAMASVAGGYY72 pKa = 10.01LGWLSSAGAQAAQAGGQARR91 pKa = 11.84IAAAAFEE98 pKa = 4.41ATRR101 pKa = 11.84AATVHH106 pKa = 6.49PAAVWANRR114 pKa = 11.84SQLVSLVASDD124 pKa = 4.62LLGFNAPAIAAVEE137 pKa = 3.85AAYY140 pKa = 10.15EE141 pKa = 4.06QMWAQDD147 pKa = 3.41VAAMFGYY154 pKa = 10.28HH155 pKa = 6.69AGASAAASALTPFIQLVQNPVASGEE180 pKa = 4.19ALVAAAQAAILSPPGRR196 pKa = 11.84VSIFNAGLADD206 pKa = 3.58VGVGNVGFGSVGDD219 pKa = 4.08GNVGAGNVGGSNVGFGNLGSLNFGSGNLGSFNVGSGNIGLYY260 pKa = 10.71NFGPGNLGSYY270 pKa = 10.35NVGLGNAGDD279 pKa = 3.91YY280 pKa = 11.39NFGFGNTGIGNIGFGNTGSHH300 pKa = 6.86NIGIGLTGDD309 pKa = 3.63GQVGFGGWNSGSANVGLFNSGIGNVGLFNSGTGNWGVGNSGEE351 pKa = 4.44LNTGLFNPGRR361 pKa = 11.84LNTGVFNTGLLNTGVGNAGSYY382 pKa = 10.96NSGSFNVGASNTGSWNAGDD401 pKa = 4.44TNTGWFNPGDD411 pKa = 3.9VNTGIGNTGDD421 pKa = 3.52VNTGGFNQGNLNNGFFWRR439 pKa = 11.84GDD441 pKa = 3.83GQGHH445 pKa = 7.23AGFDD449 pKa = 3.62YY450 pKa = 10.74TLTIPAIGLNLGADD464 pKa = 3.89VPLGIPVTGTIGTIVNGGPPEE485 pKa = 3.79ITVPGFTIPTLHH497 pKa = 6.29LTGDD501 pKa = 3.87ALFGTIGPIVVDD513 pKa = 5.16PITITGPSLNLTVGGGQSLQLGFSGPGVGPVVIPVLQVAAGPGVGNSTGVPSSGFFNSGVGGASGFGNVGGGSGWWNFGRR593 pKa = 11.84SSGVGNVGVLGSGVLNLGDD612 pKa = 4.49GVSGWFNTIPSMGSGVGNVGEE633 pKa = 4.03QLAGLFSAGPGGPSVFNLGLGNQGGLNLGHH663 pKa = 7.08ANVGDD668 pKa = 3.95FNLGGGNVGDD678 pKa = 4.6HH679 pKa = 6.15NVGGANVGDD688 pKa = 4.09ANVGVGNIGDD698 pKa = 4.11HH699 pKa = 5.98NVGGGNVGNVNVGGGNAGDD718 pKa = 4.36GNRR721 pKa = 11.84GWGNTGSFNVGFGNTGVGNFGLANQGSNNIGIGLSGDD758 pKa = 3.47NQIGFGGFNSGAGNVGLFNSGSNNIGFFNSGTGNFGVGNSGSFNTGIASTGSTNTGVFNTGLGNTGWANAGDD830 pKa = 4.11FNSGGFNAGAFNTGSFNPGDD850 pKa = 3.95TNTGWFNIGDD860 pKa = 4.44LNTGAFNTGDD870 pKa = 3.43INTGAFISGDD880 pKa = 3.49ANNGFFWRR888 pKa = 11.84GTGQGLIAADD898 pKa = 3.65YY899 pKa = 9.11TLTIPHH905 pKa = 6.92IPLTLGGGGFFEE917 pKa = 5.26LPITGSITGMTVQSFMVHH935 pKa = 7.04GIGDD939 pKa = 4.46SNTGIPLNLQLNVLGQQTDD958 pKa = 3.77EE959 pKa = 4.88IGVQVHH965 pKa = 6.14IPVIDD970 pKa = 3.8KK971 pKa = 10.8DD972 pKa = 3.87VTIPLPALPISLGITVDD989 pKa = 3.63NQIGAILVPPITINPITFNNVMVGNNTTSLSADD1022 pKa = 3.32VAGGIGPVTIPVFQLAAAPGFGNITGVPSSGFFNSGVGGGSGFGNSGSGVSGWWNVASLSSGYY1085 pKa = 10.38QNLGNLVSGFINKK1098 pKa = 9.69GEE1100 pKa = 4.1LLSGINNSNMLGLSTSGVGNVGDD1123 pKa = 3.93NVSGLFFQGADD1134 pKa = 2.57RR1135 pKa = 11.84VSIFNAGLADD1145 pKa = 3.58VGVGNVGFGSVGDD1158 pKa = 4.08GNVGAGNVGGSNVGFGNLGSLNFGSGNLGSFNVGSGNIGLYY1199 pKa = 10.71NFGPGNLGSYY1209 pKa = 10.35NVGLGNAGDD1218 pKa = 3.91YY1219 pKa = 11.39NFGFGNTGIGNIGFGNTGSHH1239 pKa = 6.86NIGIGLTGDD1248 pKa = 3.63GQVGFGGWNSGSANVGLFNSGAGNVGLFNSGTGNWGVGNSGEE1290 pKa = 4.44LNTGLFNPGRR1300 pKa = 11.84LNTGVFNTGLLNTGVGNAGSYY1321 pKa = 10.96NSGSFNVGASNTGSWNAGDD1340 pKa = 4.44TNTGWFNPGDD1350 pKa = 3.9VNTGIGNTGDD1360 pKa = 3.52VNTGGFNQGNLNNGFFWRR1378 pKa = 11.84GDD1380 pKa = 3.83GQGHH1384 pKa = 7.23AGFDD1388 pKa = 3.62YY1389 pKa = 10.74TLTIPAIALNLGVNVPLNIPITGHH1413 pKa = 6.75LGDD1416 pKa = 4.03IVIGSFTIPTLHH1428 pKa = 6.93LNGSNLTGTIGPIVVDD1444 pKa = 5.16PITITGPSLDD1454 pKa = 3.99LHH1456 pKa = 6.88VGGPGEE1462 pKa = 4.25SLQLDD1467 pKa = 3.72ISGPALGPVVIPVLQVAAGPGVGNSTGVPSSGFFNSGVGGASGFGNVGGGSGWWNFGGYY1526 pKa = 9.71SGVGNVGVLGSGVLNLGDD1544 pKa = 4.49GVSGWFNTVPSMGSGVGNVGEE1565 pKa = 4.03QLAGLFSAGPGGPSVFNLGLGNQGGLNLGHH1595 pKa = 7.08ANVGDD1600 pKa = 3.95FNLGGGNVGDD1610 pKa = 4.6HH1611 pKa = 6.15NVGGANVGDD1620 pKa = 4.09ANVGVGNIGDD1630 pKa = 4.11HH1631 pKa = 5.98NVGGGNVGNVNVGGSNAGDD1650 pKa = 3.88GNRR1653 pKa = 11.84GWGNTGSFNVGFGNTGVGNFGLANQGSNNIGIGLSGDD1690 pKa = 3.47NQIGFGGFNSGAGNVGLFNSGSNNIGFFNSGTGNFGVGNSGSFNTGIASTGSTNTGVFNTGLGNTGWANAGDD1762 pKa = 4.11FNSGGFNAGAFNTGSFNPGDD1782 pKa = 3.95TNTGWFNIGDD1792 pKa = 4.44LNTGAFNTGDD1802 pKa = 3.43INTGAFISGDD1812 pKa = 3.49ANNGFFWRR1820 pKa = 11.84GTGQGLIAADD1830 pKa = 3.65YY1831 pKa = 9.11TLTIPHH1837 pKa = 6.88IPLTLGLGGKK1847 pKa = 9.76LDD1849 pKa = 3.75IPITGQIAGLTVNPITLHH1867 pKa = 6.33GIGGSTTGIPVNAHH1881 pKa = 4.82FTLLGSTGGTTIHH1894 pKa = 6.38FNIPGTDD1901 pKa = 3.12LGFSFDD1907 pKa = 4.7LPSLPISIGVDD1918 pKa = 2.43IGDD1921 pKa = 3.76QIGAITIPQITINPITFNNVVVGGDD1946 pKa = 3.72TTSLSADD1953 pKa = 3.35VAGGIGPVTIPVFQLAAAPGFGNITGVPSSGFFNSGVGGGSGFGNSGSGVSGWWNVASLSSGYY2016 pKa = 10.38QNLGNLVSGFINKK2029 pKa = 9.69GEE2031 pKa = 4.1LLSGINNSNMLGLSTSGVGNVGDD2054 pKa = 3.93NVSGLFFQGADD2065 pKa = 2.57RR2066 pKa = 11.84VSIFNAGLADD2076 pKa = 3.58VGVGNVGFGSVGDD2089 pKa = 4.08GNVGAGNVGGSNVGFGNLGSLNFGSGNLGSFNVGSGNIGLYY2130 pKa = 10.71NFGPGNLGSYY2140 pKa = 10.35NVGLGNAGDD2149 pKa = 3.91YY2150 pKa = 11.39NFGFGNTGIGNIGFGNTGSHH2170 pKa = 6.86NIGIGLTGDD2179 pKa = 3.63GQVGFGGWNSGSANVGLFNSGAGNVGLFNSGTGNWGVGNSGEE2221 pKa = 4.44LNTGLFNPGRR2231 pKa = 11.84LNTGVFNTGLLNTGVGNAGSYY2252 pKa = 10.96NSGSFNVGASNTGSWNAGDD2271 pKa = 4.44TNTGWFNPGDD2281 pKa = 3.9VNTGIGNTGDD2291 pKa = 3.52VNTGGFNQGNLNNGFFWRR2309 pKa = 11.84GDD2311 pKa = 3.83GQGHH2315 pKa = 7.23AGFDD2319 pKa = 3.62YY2320 pKa = 10.74TLTIPAIALNLGVNVPLNIPITGHH2344 pKa = 6.63LGDD2347 pKa = 4.07IVVDD2351 pKa = 3.99PFTIPVLHH2359 pKa = 6.22LTGTGGNSLTGTIGPIVSDD2378 pKa = 3.89QITITGPSLNLTLGGPGEE2396 pKa = 4.13SLQLGFSGPALGPVVIPVLQVAAGPGVGNSTGVPSSGFFNSGVGGASGFGNVGGGSGWWNFGGYY2460 pKa = 9.71SGVGNVGVLGSGVLNLGDD2478 pKa = 4.49GVSGWFNTVPSMGSGVGNVGEE2499 pKa = 4.03QLAGLFSAGPGGPSVFNLGLGNQGGLNLGHH2529 pKa = 7.08ANVGDD2534 pKa = 3.95FNLGGGNVGDD2544 pKa = 4.6HH2545 pKa = 6.15NVGGANVGDD2554 pKa = 4.09ANVGVGNIGDD2564 pKa = 4.11HH2565 pKa = 5.98NVGGGNVGNVNVGGGNAGDD2584 pKa = 4.36GNRR2587 pKa = 11.84GWGNTGSFNVGFGNTGVGNFGLANQGSNNIGIGLSGDD2624 pKa = 3.47NQIGFGGFNSGAGNVGLFNSGSNNIGFFNSGTGNFGVGNSGSFNTGIASTGSTNTGVFNTGLGNTGWANAGDD2696 pKa = 4.11FNSGGFNAGAFNTGSFNPGDD2716 pKa = 3.95TNTGWFNIGDD2726 pKa = 4.44LNTGAFNTGDD2736 pKa = 3.43INTGAFISGDD2746 pKa = 3.49ANNGFFWRR2754 pKa = 11.84GTGQGLIAADD2764 pKa = 3.65YY2765 pKa = 9.11TLTIPHH2771 pKa = 6.88IPLTLGLGGKK2781 pKa = 9.76LDD2783 pKa = 3.75IPITGQIAGLTVNQFTLHH2801 pKa = 6.46GEE2803 pKa = 4.08NGPGIPLNLLLAIDD2817 pKa = 4.57ADD2819 pKa = 4.06SGGTSIHH2826 pKa = 6.38VGIPDD2831 pKa = 3.55TDD2833 pKa = 3.17IGFDD2837 pKa = 3.47IPIDD2841 pKa = 3.91GLPITLTLPLNSEE2854 pKa = 4.53LSPIVIEE2861 pKa = 5.08PITIGAIPLDD2871 pKa = 3.94LTVGGDD2877 pKa = 3.45TTQLGTALAAGIGPITATLFHH2898 pKa = 6.78VSPVPGFGNSTGSPSSGFFNSGAGGSSGFGNIGDD2932 pKa = 4.1TLSGFWNVGSEE2943 pKa = 4.18GSGFEE2948 pKa = 4.09NYY2950 pKa = 10.03GGSLLSGITNLGGSLSGIDD2969 pKa = 3.51NTSSLGLALAGVVSGLGNIGSQLSGLFLSGSVPP3002 pKa = 3.3
Molecular weight: 292.04 kDa
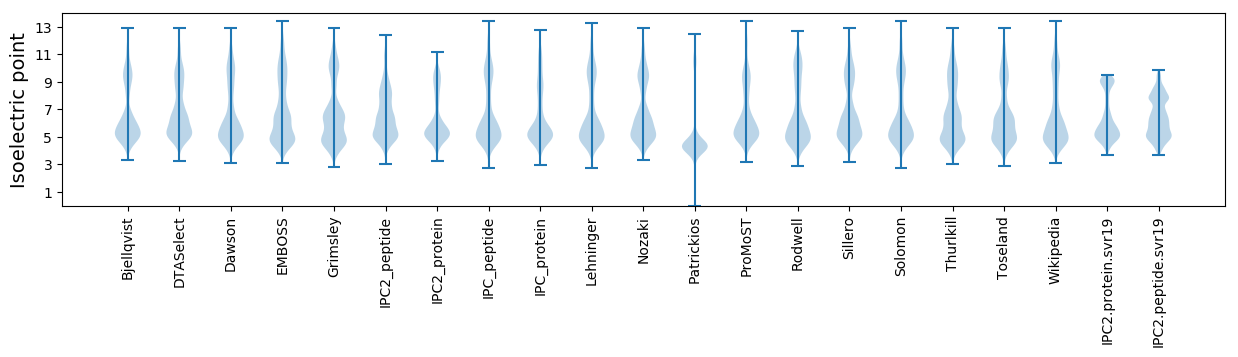
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A498QQZ4|A0A498QQZ4_9MYCO Tnp_DDE_dom domain-containing protein OS=Mycobacterium pseudokansasii OX=2341080 GN=LAUMK142_01753 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.38KK7 pKa = 8.42RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.06RR11 pKa = 11.84MSKK14 pKa = 9.76KK15 pKa = 9.54KK16 pKa = 9.72HH17 pKa = 5.63RR18 pKa = 11.84KK19 pKa = 7.56LLRR22 pKa = 11.84RR23 pKa = 11.84TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 10.11LGKK33 pKa = 9.87
MM1 pKa = 7.4GSVIKK6 pKa = 10.38KK7 pKa = 8.42RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.06RR11 pKa = 11.84MSKK14 pKa = 9.76KK15 pKa = 9.54KK16 pKa = 9.72HH17 pKa = 5.63RR18 pKa = 11.84KK19 pKa = 7.56LLRR22 pKa = 11.84RR23 pKa = 11.84TRR25 pKa = 11.84VQRR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 10.11LGKK33 pKa = 9.87
Molecular weight: 4.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1855580 |
29 |
4336 |
323.8 |
34.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.303 ± 0.052 | 0.877 ± 0.011 |
5.863 ± 0.027 | 4.772 ± 0.032 |
2.993 ± 0.022 | 9.493 ± 0.091 |
2.285 ± 0.017 | 4.174 ± 0.021 |
2.027 ± 0.022 | 9.749 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.027 ± 0.013 | 2.467 ± 0.049 |
5.975 ± 0.036 | 3.132 ± 0.017 |
7.263 ± 0.048 | 5.516 ± 0.027 |
5.974 ± 0.025 | 8.436 ± 0.036 |
1.519 ± 0.015 | 2.156 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
