
Panicum ecklonii-associated virus
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses
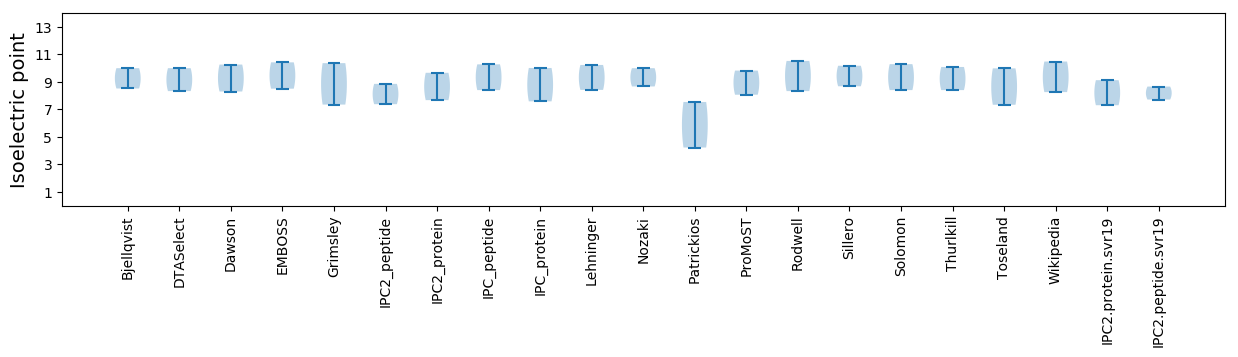
Average proteome isoelectric point is 8.21
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A345BKA6|A0A345BKA6_9VIRU Putative capsid protein OS=Panicum ecklonii-associated virus OX=2282645 PE=4 SV=1
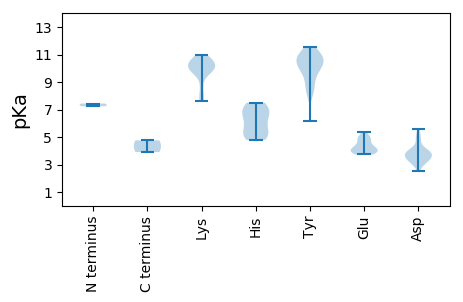
MM1 pKa = 7.44TKK3 pKa = 9.93QGSLAKK9 pKa = 9.62HH10 pKa = 4.79WVFTSFADD18 pKa = 3.47EE19 pKa = 4.24VVFFSNVDD27 pKa = 3.2VQYY30 pKa = 10.99CIFGNEE36 pKa = 4.01QCPKK40 pKa = 7.63TGRR43 pKa = 11.84WHH45 pKa = 5.3KK46 pKa = 10.32QGYY49 pKa = 8.13VVFKK53 pKa = 9.97VKK55 pKa = 10.58KK56 pKa = 10.46RR57 pKa = 11.84MTALKK62 pKa = 10.29KK63 pKa = 9.87IDD65 pKa = 3.91PTAHH69 pKa = 5.99WEE71 pKa = 3.99IKK73 pKa = 10.17RR74 pKa = 11.84GSVQQAIDD82 pKa = 3.68YY83 pKa = 8.81CKK85 pKa = 10.56KK86 pKa = 10.56DD87 pKa = 3.83DD88 pKa = 3.66DD89 pKa = 3.99WYY91 pKa = 10.48EE92 pKa = 3.95YY93 pKa = 11.26GEE95 pKa = 4.37PPVEE99 pKa = 4.06EE100 pKa = 3.88RR101 pKa = 11.84SGANAYY107 pKa = 10.34AEE109 pKa = 4.68AIAFAEE115 pKa = 4.36AGDD118 pKa = 4.17LEE120 pKa = 4.9SVKK123 pKa = 10.26QFHH126 pKa = 7.28PGTFLRR132 pKa = 11.84YY133 pKa = 9.35KK134 pKa = 9.64RR135 pKa = 11.84TLEE138 pKa = 3.8GLIKK142 pKa = 10.0YY143 pKa = 10.41DD144 pKa = 4.39CVEE147 pKa = 4.06LDD149 pKa = 3.83APCGYY154 pKa = 9.8WIYY157 pKa = 10.84GKK159 pKa = 9.66PGSGKK164 pKa = 10.07DD165 pKa = 3.59GSVMEE170 pKa = 5.01LKK172 pKa = 10.66PFVKK176 pKa = 10.01SHH178 pKa = 5.41SKK180 pKa = 8.64WWDD183 pKa = 3.42GYY185 pKa = 10.31KK186 pKa = 10.25NEE188 pKa = 4.9PYY190 pKa = 10.7VVLQDD195 pKa = 4.85LDD197 pKa = 4.89HH198 pKa = 7.5IDD200 pKa = 3.86AKK202 pKa = 10.75WIGSYY207 pKa = 10.36LKK209 pKa = 9.96QWSDD213 pKa = 3.01RR214 pKa = 11.84YY215 pKa = 11.01SFNAEE220 pKa = 3.94YY221 pKa = 10.37KK222 pKa = 9.6GGSMKK227 pKa = 9.92IRR229 pKa = 11.84PKK231 pKa = 10.44RR232 pKa = 11.84FYY234 pKa = 9.4VTSNYY239 pKa = 10.19KK240 pKa = 10.01IGEE243 pKa = 4.23LFSDD247 pKa = 5.14EE248 pKa = 3.74ICLALKK254 pKa = 10.48RR255 pKa = 11.84RR256 pKa = 11.84FHH258 pKa = 7.27VILFDD263 pKa = 3.94EE264 pKa = 5.04DD265 pKa = 3.98VVLPRR270 pKa = 11.84PVVNFVSKK278 pKa = 10.8KK279 pKa = 10.17SLIDD283 pKa = 3.36FF284 pKa = 4.76
MM1 pKa = 7.44TKK3 pKa = 9.93QGSLAKK9 pKa = 9.62HH10 pKa = 4.79WVFTSFADD18 pKa = 3.47EE19 pKa = 4.24VVFFSNVDD27 pKa = 3.2VQYY30 pKa = 10.99CIFGNEE36 pKa = 4.01QCPKK40 pKa = 7.63TGRR43 pKa = 11.84WHH45 pKa = 5.3KK46 pKa = 10.32QGYY49 pKa = 8.13VVFKK53 pKa = 9.97VKK55 pKa = 10.58KK56 pKa = 10.46RR57 pKa = 11.84MTALKK62 pKa = 10.29KK63 pKa = 9.87IDD65 pKa = 3.91PTAHH69 pKa = 5.99WEE71 pKa = 3.99IKK73 pKa = 10.17RR74 pKa = 11.84GSVQQAIDD82 pKa = 3.68YY83 pKa = 8.81CKK85 pKa = 10.56KK86 pKa = 10.56DD87 pKa = 3.83DD88 pKa = 3.66DD89 pKa = 3.99WYY91 pKa = 10.48EE92 pKa = 3.95YY93 pKa = 11.26GEE95 pKa = 4.37PPVEE99 pKa = 4.06EE100 pKa = 3.88RR101 pKa = 11.84SGANAYY107 pKa = 10.34AEE109 pKa = 4.68AIAFAEE115 pKa = 4.36AGDD118 pKa = 4.17LEE120 pKa = 4.9SVKK123 pKa = 10.26QFHH126 pKa = 7.28PGTFLRR132 pKa = 11.84YY133 pKa = 9.35KK134 pKa = 9.64RR135 pKa = 11.84TLEE138 pKa = 3.8GLIKK142 pKa = 10.0YY143 pKa = 10.41DD144 pKa = 4.39CVEE147 pKa = 4.06LDD149 pKa = 3.83APCGYY154 pKa = 9.8WIYY157 pKa = 10.84GKK159 pKa = 9.66PGSGKK164 pKa = 10.07DD165 pKa = 3.59GSVMEE170 pKa = 5.01LKK172 pKa = 10.66PFVKK176 pKa = 10.01SHH178 pKa = 5.41SKK180 pKa = 8.64WWDD183 pKa = 3.42GYY185 pKa = 10.31KK186 pKa = 10.25NEE188 pKa = 4.9PYY190 pKa = 10.7VVLQDD195 pKa = 4.85LDD197 pKa = 4.89HH198 pKa = 7.5IDD200 pKa = 3.86AKK202 pKa = 10.75WIGSYY207 pKa = 10.36LKK209 pKa = 9.96QWSDD213 pKa = 3.01RR214 pKa = 11.84YY215 pKa = 11.01SFNAEE220 pKa = 3.94YY221 pKa = 10.37KK222 pKa = 9.6GGSMKK227 pKa = 9.92IRR229 pKa = 11.84PKK231 pKa = 10.44RR232 pKa = 11.84FYY234 pKa = 9.4VTSNYY239 pKa = 10.19KK240 pKa = 10.01IGEE243 pKa = 4.23LFSDD247 pKa = 5.14EE248 pKa = 3.74ICLALKK254 pKa = 10.48RR255 pKa = 11.84RR256 pKa = 11.84FHH258 pKa = 7.27VILFDD263 pKa = 3.94EE264 pKa = 5.04DD265 pKa = 3.98VVLPRR270 pKa = 11.84PVVNFVSKK278 pKa = 10.8KK279 pKa = 10.17SLIDD283 pKa = 3.36FF284 pKa = 4.76
Molecular weight: 32.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A345BKA6|A0A345BKA6_9VIRU Putative capsid protein OS=Panicum ecklonii-associated virus OX=2282645 PE=4 SV=1
MM1 pKa = 7.24PAVRR5 pKa = 11.84RR6 pKa = 11.84LARR9 pKa = 11.84SLVRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84AVRR18 pKa = 11.84RR19 pKa = 11.84PAYY22 pKa = 9.77RR23 pKa = 11.84RR24 pKa = 11.84KK25 pKa = 9.58KK26 pKa = 7.89YY27 pKa = 8.41VRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84FHH34 pKa = 6.99RR35 pKa = 11.84KK36 pKa = 9.24RR37 pKa = 11.84FLHH40 pKa = 4.98TQNVVKK46 pKa = 7.8YY47 pKa = 9.65HH48 pKa = 6.44CKK50 pKa = 10.39QYY52 pKa = 10.01DD53 pKa = 3.38TGIMYY58 pKa = 8.85PRR60 pKa = 11.84TGSGTTFVTDD70 pKa = 3.08VQTFQLPLYY79 pKa = 10.79ANAFFNGNQQFYY91 pKa = 10.87KK92 pKa = 10.93NLDD95 pKa = 3.39EE96 pKa = 4.05YY97 pKa = 11.54HH98 pKa = 6.86FIKK101 pKa = 10.65FNYY104 pKa = 8.44IAVHH108 pKa = 5.01VKK110 pKa = 8.19EE111 pKa = 4.9LNWIGYY117 pKa = 6.17TKK119 pKa = 9.82TDD121 pKa = 2.54RR122 pKa = 11.84VVPPEE127 pKa = 4.55GGDD130 pKa = 3.46PYY132 pKa = 10.54PAVSGVTAMQMQNHH146 pKa = 6.8PVYY149 pKa = 10.77IMWDD153 pKa = 3.27IEE155 pKa = 3.85EE156 pKa = 5.36DD157 pKa = 3.91FAFDD161 pKa = 3.92TNDD164 pKa = 3.05KK165 pKa = 10.68VQITSQQLAQYY176 pKa = 9.93QGAKK180 pKa = 7.68TLRR183 pKa = 11.84TTSKK187 pKa = 10.72KK188 pKa = 8.78PVKK191 pKa = 10.03FIWRR195 pKa = 11.84FPVPWRR201 pKa = 11.84QFYY204 pKa = 10.79SCYY207 pKa = 10.12NFKK210 pKa = 10.8QITHH214 pKa = 5.25SNRR217 pKa = 11.84WGTVMEE223 pKa = 4.32EE224 pKa = 3.73LSGIKK229 pKa = 9.97NLRR232 pKa = 11.84APKK235 pKa = 9.92NLLMCHH241 pKa = 5.95PNWWGSSLPVNAALDD256 pKa = 3.75AADD259 pKa = 5.58FYY261 pKa = 11.45GQYY264 pKa = 10.83AIVYY268 pKa = 8.34HH269 pKa = 6.41LGVTFRR275 pKa = 11.84GRR277 pKa = 11.84SLMGSATSYY286 pKa = 11.1APPAQVTSADD296 pKa = 3.68EE297 pKa = 3.91
MM1 pKa = 7.24PAVRR5 pKa = 11.84RR6 pKa = 11.84LARR9 pKa = 11.84SLVRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84AVRR18 pKa = 11.84RR19 pKa = 11.84PAYY22 pKa = 9.77RR23 pKa = 11.84RR24 pKa = 11.84KK25 pKa = 9.58KK26 pKa = 7.89YY27 pKa = 8.41VRR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84FHH34 pKa = 6.99RR35 pKa = 11.84KK36 pKa = 9.24RR37 pKa = 11.84FLHH40 pKa = 4.98TQNVVKK46 pKa = 7.8YY47 pKa = 9.65HH48 pKa = 6.44CKK50 pKa = 10.39QYY52 pKa = 10.01DD53 pKa = 3.38TGIMYY58 pKa = 8.85PRR60 pKa = 11.84TGSGTTFVTDD70 pKa = 3.08VQTFQLPLYY79 pKa = 10.79ANAFFNGNQQFYY91 pKa = 10.87KK92 pKa = 10.93NLDD95 pKa = 3.39EE96 pKa = 4.05YY97 pKa = 11.54HH98 pKa = 6.86FIKK101 pKa = 10.65FNYY104 pKa = 8.44IAVHH108 pKa = 5.01VKK110 pKa = 8.19EE111 pKa = 4.9LNWIGYY117 pKa = 6.17TKK119 pKa = 9.82TDD121 pKa = 2.54RR122 pKa = 11.84VVPPEE127 pKa = 4.55GGDD130 pKa = 3.46PYY132 pKa = 10.54PAVSGVTAMQMQNHH146 pKa = 6.8PVYY149 pKa = 10.77IMWDD153 pKa = 3.27IEE155 pKa = 3.85EE156 pKa = 5.36DD157 pKa = 3.91FAFDD161 pKa = 3.92TNDD164 pKa = 3.05KK165 pKa = 10.68VQITSQQLAQYY176 pKa = 9.93QGAKK180 pKa = 7.68TLRR183 pKa = 11.84TTSKK187 pKa = 10.72KK188 pKa = 8.78PVKK191 pKa = 10.03FIWRR195 pKa = 11.84FPVPWRR201 pKa = 11.84QFYY204 pKa = 10.79SCYY207 pKa = 10.12NFKK210 pKa = 10.8QITHH214 pKa = 5.25SNRR217 pKa = 11.84WGTVMEE223 pKa = 4.32EE224 pKa = 3.73LSGIKK229 pKa = 9.97NLRR232 pKa = 11.84APKK235 pKa = 9.92NLLMCHH241 pKa = 5.95PNWWGSSLPVNAALDD256 pKa = 3.75AADD259 pKa = 5.58FYY261 pKa = 11.45GQYY264 pKa = 10.83AIVYY268 pKa = 8.34HH269 pKa = 6.41LGVTFRR275 pKa = 11.84GRR277 pKa = 11.84SLMGSATSYY286 pKa = 11.1APPAQVTSADD296 pKa = 3.68EE297 pKa = 3.91
Molecular weight: 34.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
581 |
284 |
297 |
290.5 |
33.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.54 ± 0.658 | 1.549 ± 0.409 |
5.508 ± 1.113 | 4.475 ± 1.352 |
5.852 ± 0.097 | 6.196 ± 0.614 |
2.754 ± 0.21 | 4.303 ± 0.455 |
8.09 ± 1.795 | 5.852 ± 0.097 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.065 ± 0.477 | 3.787 ± 0.959 |
4.991 ± 0.556 | 4.475 ± 0.948 |
6.368 ± 1.555 | 5.508 ± 0.602 |
4.819 ± 1.453 | 7.917 ± 0.131 |
2.754 ± 0.301 | 6.196 ± 0.153 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
