
Human papillomavirus 93
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Betapapillomavirus; Betapapillomavirus 1
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
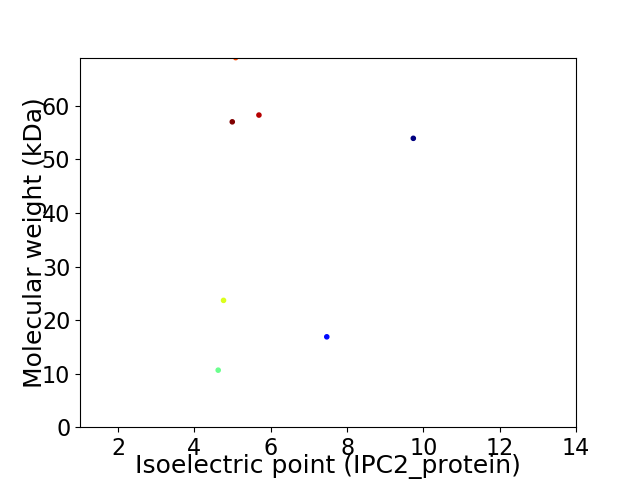
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q6TY36|Q6TY36_9PAPI Protein E6 OS=Human papillomavirus 93 OX=247268 GN=E6 PE=3 SV=1
MM1 pKa = 7.37IGKK4 pKa = 8.93EE5 pKa = 3.9VTLQDD10 pKa = 3.25IVLGLTPPEE19 pKa = 3.95PVALNCEE26 pKa = 4.31EE27 pKa = 4.39EE28 pKa = 4.91LPTEE32 pKa = 4.0QDD34 pKa = 3.37TEE36 pKa = 4.24EE37 pKa = 4.38EE38 pKa = 4.17PARR41 pKa = 11.84TAFKK45 pKa = 10.46IIVFCGGGCGSRR57 pKa = 11.84LRR59 pKa = 11.84IFVAATEE66 pKa = 4.15FGVRR70 pKa = 11.84CFQQLLVKK78 pKa = 10.2EE79 pKa = 4.73LDD81 pKa = 4.01LLCPNCRR88 pKa = 11.84NSDD91 pKa = 3.87LQHH94 pKa = 6.61GGQQ97 pKa = 3.6
MM1 pKa = 7.37IGKK4 pKa = 8.93EE5 pKa = 3.9VTLQDD10 pKa = 3.25IVLGLTPPEE19 pKa = 3.95PVALNCEE26 pKa = 4.31EE27 pKa = 4.39EE28 pKa = 4.91LPTEE32 pKa = 4.0QDD34 pKa = 3.37TEE36 pKa = 4.24EE37 pKa = 4.38EE38 pKa = 4.17PARR41 pKa = 11.84TAFKK45 pKa = 10.46IIVFCGGGCGSRR57 pKa = 11.84LRR59 pKa = 11.84IFVAATEE66 pKa = 4.15FGVRR70 pKa = 11.84CFQQLLVKK78 pKa = 10.2EE79 pKa = 4.73LDD81 pKa = 4.01LLCPNCRR88 pKa = 11.84NSDD91 pKa = 3.87LQHH94 pKa = 6.61GGQQ97 pKa = 3.6
Molecular weight: 10.65 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q6TY34|Q6TY34_9PAPI Replication protein E1 OS=Human papillomavirus 93 OX=247268 GN=E1 PE=3 SV=1
MM1 pKa = 6.59EE2 pKa = 4.56TLRR5 pKa = 11.84EE6 pKa = 4.06RR7 pKa = 11.84FDD9 pKa = 3.56VLQDD13 pKa = 3.26QLMNIYY19 pKa = 9.69EE20 pKa = 4.36VASDD24 pKa = 3.5TLEE27 pKa = 3.89AQIEE31 pKa = 4.24HH32 pKa = 6.31WRR34 pKa = 11.84LLRR37 pKa = 11.84KK38 pKa = 9.23EE39 pKa = 4.18AVLLYY44 pKa = 8.99FARR47 pKa = 11.84QNGVLRR53 pKa = 11.84LGYY56 pKa = 10.31QPVPPLATSEE66 pKa = 4.35AKK68 pKa = 10.45AKK70 pKa = 9.92EE71 pKa = 4.49AIGMMLQLQSLQKK84 pKa = 10.19SAYY87 pKa = 9.84ASEE90 pKa = 3.96KK91 pKa = 7.62WTLVDD96 pKa = 3.78TSIEE100 pKa = 4.32TFKK103 pKa = 10.35NTPEE107 pKa = 3.73NHH109 pKa = 6.11FKK111 pKa = 10.73KK112 pKa = 10.7GPVNVEE118 pKa = 3.83VIYY121 pKa = 11.2DD122 pKa = 4.14GDD124 pKa = 4.07PDD126 pKa = 3.81NANLYY131 pKa = 8.07TMWKK135 pKa = 7.81YY136 pKa = 11.35VYY138 pKa = 10.55YY139 pKa = 10.55LDD141 pKa = 5.93SEE143 pKa = 4.99DD144 pKa = 3.7RR145 pKa = 11.84WQKK148 pKa = 9.18TEE150 pKa = 3.57SGANHH155 pKa = 6.03TGIYY159 pKa = 8.44YY160 pKa = 8.7TIKK163 pKa = 10.57DD164 pKa = 3.77FKK166 pKa = 10.91HH167 pKa = 6.43YY168 pKa = 8.5YY169 pKa = 9.45TLFADD174 pKa = 4.09DD175 pKa = 3.56AKK177 pKa = 11.05KK178 pKa = 10.63YY179 pKa = 10.05GKK181 pKa = 9.88SGQWEE186 pKa = 3.89VRR188 pKa = 11.84INKK191 pKa = 7.16EE192 pKa = 4.11TVFAPVTSSTPPEE205 pKa = 4.08SPGRR209 pKa = 11.84EE210 pKa = 3.53RR211 pKa = 11.84ASPEE215 pKa = 3.92STAHH219 pKa = 5.85TKK221 pKa = 7.25TTARR225 pKa = 11.84SSASPDD231 pKa = 3.23HH232 pKa = 6.67EE233 pKa = 5.22LPQQTSDD240 pKa = 2.87EE241 pKa = 4.57TNRR244 pKa = 11.84KK245 pKa = 8.2RR246 pKa = 11.84RR247 pKa = 11.84YY248 pKa = 9.09GRR250 pKa = 11.84RR251 pKa = 11.84EE252 pKa = 3.74SSPTEE257 pKa = 3.95SKK259 pKa = 10.29RR260 pKa = 11.84KK261 pKa = 9.19RR262 pKa = 11.84RR263 pKa = 11.84RR264 pKa = 11.84SSSRR268 pKa = 11.84QKK270 pKa = 9.84KK271 pKa = 7.6QGRR274 pKa = 11.84RR275 pKa = 11.84SRR277 pKa = 11.84SRR279 pKa = 11.84TTSSQSRR286 pKa = 11.84SRR288 pKa = 11.84SRR290 pKa = 11.84SSSSRR295 pKa = 11.84GSRR298 pKa = 11.84GSRR301 pKa = 11.84GRR303 pKa = 11.84TPRR306 pKa = 11.84GRR308 pKa = 11.84GKK310 pKa = 8.46HH311 pKa = 3.89TTRR314 pKa = 11.84GRR316 pKa = 11.84GRR318 pKa = 11.84GSGRR322 pKa = 11.84KK323 pKa = 8.41GDD325 pKa = 3.37RR326 pKa = 11.84RR327 pKa = 11.84GRR329 pKa = 11.84RR330 pKa = 11.84SRR332 pKa = 11.84RR333 pKa = 11.84SSSSSSPTICTRR345 pKa = 11.84SASQTRR351 pKa = 11.84SKK353 pKa = 10.69QSKK356 pKa = 8.05CARR359 pKa = 11.84DD360 pKa = 3.79GGISPGDD367 pKa = 3.45VGRR370 pKa = 11.84SVQTVSGRR378 pKa = 11.84NTGRR382 pKa = 11.84LGRR385 pKa = 11.84LLEE388 pKa = 4.17EE389 pKa = 4.57ARR391 pKa = 11.84DD392 pKa = 3.73PPVILIRR399 pKa = 11.84GEE401 pKa = 4.09ANTVKK406 pKa = 10.54CFRR409 pKa = 11.84NRR411 pKa = 11.84AKK413 pKa = 10.32QKK415 pKa = 11.09YY416 pKa = 9.3KK417 pKa = 10.64GLCKK421 pKa = 10.39AFSTTWSWVAADD433 pKa = 3.41GTEE436 pKa = 3.7RR437 pKa = 11.84LGRR440 pKa = 11.84SRR442 pKa = 11.84VLVSFTSHH450 pKa = 5.1TQRR453 pKa = 11.84SSFLKK458 pKa = 10.05VVKK461 pKa = 10.2FPKK464 pKa = 10.48GVDD467 pKa = 3.05WSLGNLDD474 pKa = 4.16KK475 pKa = 11.47LL476 pKa = 4.08
MM1 pKa = 6.59EE2 pKa = 4.56TLRR5 pKa = 11.84EE6 pKa = 4.06RR7 pKa = 11.84FDD9 pKa = 3.56VLQDD13 pKa = 3.26QLMNIYY19 pKa = 9.69EE20 pKa = 4.36VASDD24 pKa = 3.5TLEE27 pKa = 3.89AQIEE31 pKa = 4.24HH32 pKa = 6.31WRR34 pKa = 11.84LLRR37 pKa = 11.84KK38 pKa = 9.23EE39 pKa = 4.18AVLLYY44 pKa = 8.99FARR47 pKa = 11.84QNGVLRR53 pKa = 11.84LGYY56 pKa = 10.31QPVPPLATSEE66 pKa = 4.35AKK68 pKa = 10.45AKK70 pKa = 9.92EE71 pKa = 4.49AIGMMLQLQSLQKK84 pKa = 10.19SAYY87 pKa = 9.84ASEE90 pKa = 3.96KK91 pKa = 7.62WTLVDD96 pKa = 3.78TSIEE100 pKa = 4.32TFKK103 pKa = 10.35NTPEE107 pKa = 3.73NHH109 pKa = 6.11FKK111 pKa = 10.73KK112 pKa = 10.7GPVNVEE118 pKa = 3.83VIYY121 pKa = 11.2DD122 pKa = 4.14GDD124 pKa = 4.07PDD126 pKa = 3.81NANLYY131 pKa = 8.07TMWKK135 pKa = 7.81YY136 pKa = 11.35VYY138 pKa = 10.55YY139 pKa = 10.55LDD141 pKa = 5.93SEE143 pKa = 4.99DD144 pKa = 3.7RR145 pKa = 11.84WQKK148 pKa = 9.18TEE150 pKa = 3.57SGANHH155 pKa = 6.03TGIYY159 pKa = 8.44YY160 pKa = 8.7TIKK163 pKa = 10.57DD164 pKa = 3.77FKK166 pKa = 10.91HH167 pKa = 6.43YY168 pKa = 8.5YY169 pKa = 9.45TLFADD174 pKa = 4.09DD175 pKa = 3.56AKK177 pKa = 11.05KK178 pKa = 10.63YY179 pKa = 10.05GKK181 pKa = 9.88SGQWEE186 pKa = 3.89VRR188 pKa = 11.84INKK191 pKa = 7.16EE192 pKa = 4.11TVFAPVTSSTPPEE205 pKa = 4.08SPGRR209 pKa = 11.84EE210 pKa = 3.53RR211 pKa = 11.84ASPEE215 pKa = 3.92STAHH219 pKa = 5.85TKK221 pKa = 7.25TTARR225 pKa = 11.84SSASPDD231 pKa = 3.23HH232 pKa = 6.67EE233 pKa = 5.22LPQQTSDD240 pKa = 2.87EE241 pKa = 4.57TNRR244 pKa = 11.84KK245 pKa = 8.2RR246 pKa = 11.84RR247 pKa = 11.84YY248 pKa = 9.09GRR250 pKa = 11.84RR251 pKa = 11.84EE252 pKa = 3.74SSPTEE257 pKa = 3.95SKK259 pKa = 10.29RR260 pKa = 11.84KK261 pKa = 9.19RR262 pKa = 11.84RR263 pKa = 11.84RR264 pKa = 11.84SSSRR268 pKa = 11.84QKK270 pKa = 9.84KK271 pKa = 7.6QGRR274 pKa = 11.84RR275 pKa = 11.84SRR277 pKa = 11.84SRR279 pKa = 11.84TTSSQSRR286 pKa = 11.84SRR288 pKa = 11.84SRR290 pKa = 11.84SSSSRR295 pKa = 11.84GSRR298 pKa = 11.84GSRR301 pKa = 11.84GRR303 pKa = 11.84TPRR306 pKa = 11.84GRR308 pKa = 11.84GKK310 pKa = 8.46HH311 pKa = 3.89TTRR314 pKa = 11.84GRR316 pKa = 11.84GRR318 pKa = 11.84GSGRR322 pKa = 11.84KK323 pKa = 8.41GDD325 pKa = 3.37RR326 pKa = 11.84RR327 pKa = 11.84GRR329 pKa = 11.84RR330 pKa = 11.84SRR332 pKa = 11.84RR333 pKa = 11.84SSSSSSPTICTRR345 pKa = 11.84SASQTRR351 pKa = 11.84SKK353 pKa = 10.69QSKK356 pKa = 8.05CARR359 pKa = 11.84DD360 pKa = 3.79GGISPGDD367 pKa = 3.45VGRR370 pKa = 11.84SVQTVSGRR378 pKa = 11.84NTGRR382 pKa = 11.84LGRR385 pKa = 11.84LLEE388 pKa = 4.17EE389 pKa = 4.57ARR391 pKa = 11.84DD392 pKa = 3.73PPVILIRR399 pKa = 11.84GEE401 pKa = 4.09ANTVKK406 pKa = 10.54CFRR409 pKa = 11.84NRR411 pKa = 11.84AKK413 pKa = 10.32QKK415 pKa = 11.09YY416 pKa = 9.3KK417 pKa = 10.64GLCKK421 pKa = 10.39AFSTTWSWVAADD433 pKa = 3.41GTEE436 pKa = 3.7RR437 pKa = 11.84LGRR440 pKa = 11.84SRR442 pKa = 11.84VLVSFTSHH450 pKa = 5.1TQRR453 pKa = 11.84SSFLKK458 pKa = 10.05VVKK461 pKa = 10.2FPKK464 pKa = 10.48GVDD467 pKa = 3.05WSLGNLDD474 pKa = 4.16KK475 pKa = 11.47LL476 pKa = 4.08
Molecular weight: 53.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2572 |
97 |
603 |
367.4 |
41.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.404 ± 0.371 | 1.944 ± 0.827 |
5.793 ± 0.397 | 6.882 ± 0.82 |
3.771 ± 0.54 | 6.61 ± 0.744 |
1.75 ± 0.3 | 4.238 ± 0.574 |
5.21 ± 0.853 | 8.826 ± 0.972 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.439 ± 0.417 | 4.471 ± 0.866 |
6.415 ± 1.315 | 4.977 ± 0.299 |
6.882 ± 1.47 | 8.32 ± 1.251 |
6.26 ± 0.721 | 6.182 ± 0.638 |
1.283 ± 0.347 | 3.344 ± 0.491 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
