
Faeces associated gemycircularvirus 11
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemykroznavirus; Gemykroznavirus rabas1; Rabbit associated gemykroznavirus 1
Average proteome isoelectric point is 7.27
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
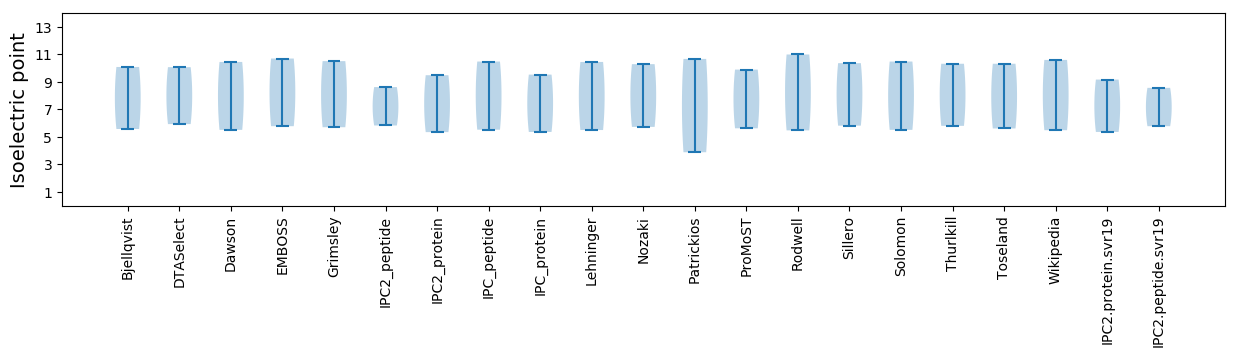
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|T1YTA5|T1YTA5_9VIRU Replication associated protein OS=Faeces associated gemycircularvirus 11 OX=1391026 PE=3 SV=1
MM1 pKa = 7.28LVARR5 pKa = 11.84SSVPPRR11 pKa = 11.84VMPRR15 pKa = 11.84TVNWSVALEE24 pKa = 4.08SVAFLTYY31 pKa = 10.22SQYY34 pKa = 11.4GPADD38 pKa = 3.63GVSDD42 pKa = 3.67IQASSQCFEE51 pKa = 4.33YY52 pKa = 10.33IIILVAHH59 pKa = 6.59EE60 pKa = 3.85RR61 pKa = 11.84HH62 pKa = 6.34EE63 pKa = 5.04DD64 pKa = 3.63GGHH67 pKa = 6.33HH68 pKa = 5.23YY69 pKa = 10.14HH70 pKa = 7.05VLVAWEE76 pKa = 4.33SHH78 pKa = 5.41FKK80 pKa = 11.21ANDD83 pKa = 2.96VRR85 pKa = 11.84IFDD88 pKa = 4.71VGGCHH93 pKa = 7.07PNFKK97 pKa = 10.26SVRR100 pKa = 11.84GDD102 pKa = 3.39GNINRR107 pKa = 11.84VLEE110 pKa = 4.06QLRR113 pKa = 11.84LAQFLPGYY121 pKa = 8.35CLKK124 pKa = 11.18DD125 pKa = 2.95GDD127 pKa = 4.57YY128 pKa = 11.31YY129 pKa = 11.48GDD131 pKa = 3.6PEE133 pKa = 4.49VFRR136 pKa = 11.84RR137 pKa = 11.84DD138 pKa = 3.29APKK141 pKa = 10.43RR142 pKa = 11.84SQTSIWTEE150 pKa = 3.66IVSAEE155 pKa = 4.09SAGQFLQLARR165 pKa = 11.84EE166 pKa = 4.41LAPKK170 pKa = 10.16EE171 pKa = 3.68YY172 pKa = 10.49GLNYY176 pKa = 10.01DD177 pKa = 4.39RR178 pKa = 11.84LVSYY182 pKa = 10.69CRR184 pKa = 11.84AHH186 pKa = 6.0YY187 pKa = 8.54DD188 pKa = 3.09KK189 pKa = 10.6PAEE192 pKa = 4.01YY193 pKa = 10.53VPRR196 pKa = 11.84HH197 pKa = 5.32VDD199 pKa = 3.7FVVPSEE205 pKa = 3.95LDD207 pKa = 2.9DD208 pKa = 3.72WVNVNVSNAAPEE220 pKa = 4.11QPEE223 pKa = 4.22RR224 pKa = 11.84PRR226 pKa = 11.84ALCLIGRR233 pKa = 11.84SRR235 pKa = 11.84LGKK238 pKa = 7.97TEE240 pKa = 3.82WARR243 pKa = 11.84SLGKK247 pKa = 9.77HH248 pKa = 5.98SYY250 pKa = 9.45FASLFNIDD258 pKa = 5.24DD259 pKa = 4.05FDD261 pKa = 5.38GSADD265 pKa = 3.76YY266 pKa = 11.48AVMDD270 pKa = 5.72DD271 pKa = 3.68IDD273 pKa = 4.32IKK275 pKa = 10.89FFPNYY280 pKa = 9.54KK281 pKa = 9.9GWFGAQKK288 pKa = 10.17RR289 pKa = 11.84FCITGKK295 pKa = 8.46YY296 pKa = 8.94ARR298 pKa = 11.84HH299 pKa = 5.49RR300 pKa = 11.84TVDD303 pKa = 2.85WGKK306 pKa = 10.08PMIWCTNDD314 pKa = 3.66DD315 pKa = 3.4PRR317 pKa = 11.84QAVGIDD323 pKa = 3.29QDD325 pKa = 3.81WFSEE329 pKa = 4.14NPDD332 pKa = 3.35FVIVQEE338 pKa = 4.07PFLLGSLQVALALATDD354 pKa = 4.06SPRR357 pKa = 11.84FDD359 pKa = 3.36SS360 pKa = 4.38
MM1 pKa = 7.28LVARR5 pKa = 11.84SSVPPRR11 pKa = 11.84VMPRR15 pKa = 11.84TVNWSVALEE24 pKa = 4.08SVAFLTYY31 pKa = 10.22SQYY34 pKa = 11.4GPADD38 pKa = 3.63GVSDD42 pKa = 3.67IQASSQCFEE51 pKa = 4.33YY52 pKa = 10.33IIILVAHH59 pKa = 6.59EE60 pKa = 3.85RR61 pKa = 11.84HH62 pKa = 6.34EE63 pKa = 5.04DD64 pKa = 3.63GGHH67 pKa = 6.33HH68 pKa = 5.23YY69 pKa = 10.14HH70 pKa = 7.05VLVAWEE76 pKa = 4.33SHH78 pKa = 5.41FKK80 pKa = 11.21ANDD83 pKa = 2.96VRR85 pKa = 11.84IFDD88 pKa = 4.71VGGCHH93 pKa = 7.07PNFKK97 pKa = 10.26SVRR100 pKa = 11.84GDD102 pKa = 3.39GNINRR107 pKa = 11.84VLEE110 pKa = 4.06QLRR113 pKa = 11.84LAQFLPGYY121 pKa = 8.35CLKK124 pKa = 11.18DD125 pKa = 2.95GDD127 pKa = 4.57YY128 pKa = 11.31YY129 pKa = 11.48GDD131 pKa = 3.6PEE133 pKa = 4.49VFRR136 pKa = 11.84RR137 pKa = 11.84DD138 pKa = 3.29APKK141 pKa = 10.43RR142 pKa = 11.84SQTSIWTEE150 pKa = 3.66IVSAEE155 pKa = 4.09SAGQFLQLARR165 pKa = 11.84EE166 pKa = 4.41LAPKK170 pKa = 10.16EE171 pKa = 3.68YY172 pKa = 10.49GLNYY176 pKa = 10.01DD177 pKa = 4.39RR178 pKa = 11.84LVSYY182 pKa = 10.69CRR184 pKa = 11.84AHH186 pKa = 6.0YY187 pKa = 8.54DD188 pKa = 3.09KK189 pKa = 10.6PAEE192 pKa = 4.01YY193 pKa = 10.53VPRR196 pKa = 11.84HH197 pKa = 5.32VDD199 pKa = 3.7FVVPSEE205 pKa = 3.95LDD207 pKa = 2.9DD208 pKa = 3.72WVNVNVSNAAPEE220 pKa = 4.11QPEE223 pKa = 4.22RR224 pKa = 11.84PRR226 pKa = 11.84ALCLIGRR233 pKa = 11.84SRR235 pKa = 11.84LGKK238 pKa = 7.97TEE240 pKa = 3.82WARR243 pKa = 11.84SLGKK247 pKa = 9.77HH248 pKa = 5.98SYY250 pKa = 9.45FASLFNIDD258 pKa = 5.24DD259 pKa = 4.05FDD261 pKa = 5.38GSADD265 pKa = 3.76YY266 pKa = 11.48AVMDD270 pKa = 5.72DD271 pKa = 3.68IDD273 pKa = 4.32IKK275 pKa = 10.89FFPNYY280 pKa = 9.54KK281 pKa = 9.9GWFGAQKK288 pKa = 10.17RR289 pKa = 11.84FCITGKK295 pKa = 8.46YY296 pKa = 8.94ARR298 pKa = 11.84HH299 pKa = 5.49RR300 pKa = 11.84TVDD303 pKa = 2.85WGKK306 pKa = 10.08PMIWCTNDD314 pKa = 3.66DD315 pKa = 3.4PRR317 pKa = 11.84QAVGIDD323 pKa = 3.29QDD325 pKa = 3.81WFSEE329 pKa = 4.14NPDD332 pKa = 3.35FVIVQEE338 pKa = 4.07PFLLGSLQVALALATDD354 pKa = 4.06SPRR357 pKa = 11.84FDD359 pKa = 3.36SS360 pKa = 4.38
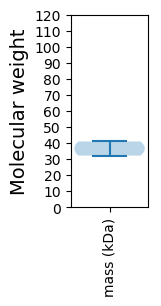
Molecular weight: 40.94 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|T1YTA5|T1YTA5_9VIRU Replication associated protein OS=Faeces associated gemycircularvirus 11 OX=1391026 PE=3 SV=1
MM1 pKa = 7.47PRR3 pKa = 11.84YY4 pKa = 9.69AKK6 pKa = 9.26RR7 pKa = 11.84TSKK10 pKa = 10.26KK11 pKa = 9.54RR12 pKa = 11.84PAYY15 pKa = 9.56RR16 pKa = 11.84KK17 pKa = 8.54KK18 pKa = 10.17AAYY21 pKa = 9.46RR22 pKa = 11.84GKK24 pKa = 10.41KK25 pKa = 8.02RR26 pKa = 11.84TYY28 pKa = 9.94SKK30 pKa = 9.66KK31 pKa = 9.03TSSSLAKK38 pKa = 10.01KK39 pKa = 9.2IHH41 pKa = 6.18AVADD45 pKa = 4.04TKK47 pKa = 11.23KK48 pKa = 10.45SDD50 pKa = 3.3TMACAIAAPVTGANPSGVEE69 pKa = 3.96VKK71 pKa = 10.73EE72 pKa = 4.19FMDD75 pKa = 3.96WPLDD79 pKa = 3.9SQIKK83 pKa = 9.58IYY85 pKa = 9.3MWAPLYY91 pKa = 10.65RR92 pKa = 11.84VGTDD96 pKa = 2.54KK97 pKa = 11.08KK98 pKa = 11.04GPHH101 pKa = 6.39LRR103 pKa = 11.84NASRR107 pKa = 11.84VHH109 pKa = 4.41FTGVSEE115 pKa = 4.38RR116 pKa = 11.84MNIASNSRR124 pKa = 11.84VPFRR128 pKa = 11.84HH129 pKa = 5.44RR130 pKa = 11.84RR131 pKa = 11.84LVIRR135 pKa = 11.84AAVDD139 pKa = 3.69FTSRR143 pKa = 11.84MLASKK148 pKa = 10.74ADD150 pKa = 3.53PTLVFRR156 pKa = 11.84NLDD159 pKa = 3.48QLTPTALDD167 pKa = 3.78TYY169 pKa = 11.26FLGQKK174 pKa = 9.66NLDD177 pKa = 3.15WSDD180 pKa = 4.13PIKK183 pKa = 11.09ADD185 pKa = 3.29VDD187 pKa = 3.89KK188 pKa = 11.41QQLTVLFDD196 pKa = 3.05KK197 pKa = 10.63TYY199 pKa = 10.37TYY201 pKa = 11.22NPPNDD206 pKa = 3.85FGLDD210 pKa = 3.51STRR213 pKa = 11.84TSITRR218 pKa = 11.84LTAKK222 pKa = 10.33VGLPTRR228 pKa = 11.84KK229 pKa = 10.21DD230 pKa = 3.38GVNTKK235 pKa = 8.51WSAKK239 pKa = 9.99IGTKK243 pKa = 9.9EE244 pKa = 3.96PNDD247 pKa = 3.51VVIIDD252 pKa = 4.16MFRR255 pKa = 11.84SNYY258 pKa = 7.46DD259 pKa = 2.8TRR261 pKa = 11.84TNTFPLGIEE270 pKa = 3.81SWTIGSKK277 pKa = 10.52CKK279 pKa = 10.45GYY281 pKa = 11.14LEE283 pKa = 4.49GTT285 pKa = 3.83
MM1 pKa = 7.47PRR3 pKa = 11.84YY4 pKa = 9.69AKK6 pKa = 9.26RR7 pKa = 11.84TSKK10 pKa = 10.26KK11 pKa = 9.54RR12 pKa = 11.84PAYY15 pKa = 9.56RR16 pKa = 11.84KK17 pKa = 8.54KK18 pKa = 10.17AAYY21 pKa = 9.46RR22 pKa = 11.84GKK24 pKa = 10.41KK25 pKa = 8.02RR26 pKa = 11.84TYY28 pKa = 9.94SKK30 pKa = 9.66KK31 pKa = 9.03TSSSLAKK38 pKa = 10.01KK39 pKa = 9.2IHH41 pKa = 6.18AVADD45 pKa = 4.04TKK47 pKa = 11.23KK48 pKa = 10.45SDD50 pKa = 3.3TMACAIAAPVTGANPSGVEE69 pKa = 3.96VKK71 pKa = 10.73EE72 pKa = 4.19FMDD75 pKa = 3.96WPLDD79 pKa = 3.9SQIKK83 pKa = 9.58IYY85 pKa = 9.3MWAPLYY91 pKa = 10.65RR92 pKa = 11.84VGTDD96 pKa = 2.54KK97 pKa = 11.08KK98 pKa = 11.04GPHH101 pKa = 6.39LRR103 pKa = 11.84NASRR107 pKa = 11.84VHH109 pKa = 4.41FTGVSEE115 pKa = 4.38RR116 pKa = 11.84MNIASNSRR124 pKa = 11.84VPFRR128 pKa = 11.84HH129 pKa = 5.44RR130 pKa = 11.84RR131 pKa = 11.84LVIRR135 pKa = 11.84AAVDD139 pKa = 3.69FTSRR143 pKa = 11.84MLASKK148 pKa = 10.74ADD150 pKa = 3.53PTLVFRR156 pKa = 11.84NLDD159 pKa = 3.48QLTPTALDD167 pKa = 3.78TYY169 pKa = 11.26FLGQKK174 pKa = 9.66NLDD177 pKa = 3.15WSDD180 pKa = 4.13PIKK183 pKa = 11.09ADD185 pKa = 3.29VDD187 pKa = 3.89KK188 pKa = 11.41QQLTVLFDD196 pKa = 3.05KK197 pKa = 10.63TYY199 pKa = 10.37TYY201 pKa = 11.22NPPNDD206 pKa = 3.85FGLDD210 pKa = 3.51STRR213 pKa = 11.84TSITRR218 pKa = 11.84LTAKK222 pKa = 10.33VGLPTRR228 pKa = 11.84KK229 pKa = 10.21DD230 pKa = 3.38GVNTKK235 pKa = 8.51WSAKK239 pKa = 9.99IGTKK243 pKa = 9.9EE244 pKa = 3.96PNDD247 pKa = 3.51VVIIDD252 pKa = 4.16MFRR255 pKa = 11.84SNYY258 pKa = 7.46DD259 pKa = 2.8TRR261 pKa = 11.84TNTFPLGIEE270 pKa = 3.81SWTIGSKK277 pKa = 10.52CKK279 pKa = 10.45GYY281 pKa = 11.14LEE283 pKa = 4.49GTT285 pKa = 3.83
Molecular weight: 32.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
645 |
285 |
360 |
322.5 |
36.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.062 ± 0.005 | 1.395 ± 0.427 |
7.752 ± 0.452 | 3.721 ± 0.995 |
4.651 ± 0.703 | 5.891 ± 0.387 |
2.326 ± 0.568 | 4.496 ± 0.04 |
6.512 ± 2.256 | 6.977 ± 0.191 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.705 ± 0.462 | 3.876 ± 0.206 |
5.736 ± 0.075 | 2.946 ± 0.734 |
7.287 ± 0.266 | 7.287 ± 0.05 |
5.736 ± 2.517 | 7.287 ± 0.814 |
2.171 ± 0.256 | 4.186 ± 0.201 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
