
Deinococcus sp. HMF7620
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Deinococcus-Thermus; Deinococci; Deinococcales; Deinococcaceae; Deinococcus; unclassified Deinococcus
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
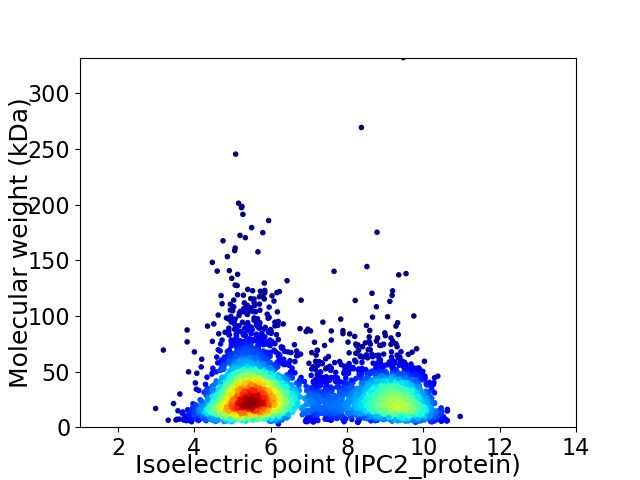
Virtual 2D-PAGE plot for 4347 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A7C9HU66|A0A7C9HU66_9DEIO Phospholipase OS=Deinococcus sp. HMF7620 OX=2682977 GN=GO986_20555 PE=4 SV=1
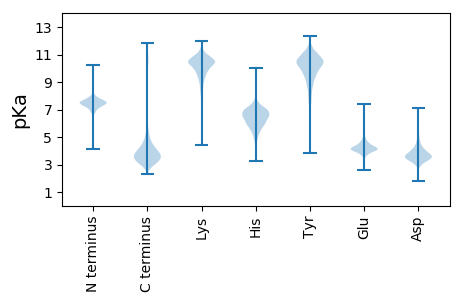
MM1 pKa = 7.27SRR3 pKa = 11.84LSVRR7 pKa = 11.84LLCTVVGLSLLSSALAVGTPVNTSISNTATLNYY40 pKa = 9.69RR41 pKa = 11.84DD42 pKa = 4.47ALNAPRR48 pKa = 11.84TQTSNPVTVTVRR60 pKa = 11.84QVYY63 pKa = 10.07ALSLTPDD70 pKa = 3.46AAEE73 pKa = 3.88NAIPLSRR80 pKa = 11.84QFSAQAGASRR90 pKa = 11.84LIGYY94 pKa = 7.4EE95 pKa = 3.93LVNAGNGPDD104 pKa = 3.71TLTLGVTQSATDD116 pKa = 3.73GFDD119 pKa = 2.85VTTRR123 pKa = 11.84LLPDD127 pKa = 4.29ANCDD131 pKa = 3.46GTVDD135 pKa = 3.66SAVPLGAAQTLAADD149 pKa = 3.95EE150 pKa = 4.36RR151 pKa = 11.84LCVYY155 pKa = 10.53VEE157 pKa = 3.97ATVPAGAPTGAAARR171 pKa = 11.84LTLTAASQGNPAVTDD186 pKa = 3.68TEE188 pKa = 4.79NYY190 pKa = 9.9AQVTVSAAGQLDD202 pKa = 3.38ITKK205 pKa = 8.65TVSPTGSALPGAPLTYY221 pKa = 9.1TVSGSVPAGNPVGAVSGVLTVDD243 pKa = 3.93GVPRR247 pKa = 11.84SGVLVRR253 pKa = 11.84DD254 pKa = 3.59VLTTLEE260 pKa = 4.37FGAVTAASASSGAATALYY278 pKa = 8.97STDD281 pKa = 5.22GGATWTAAVPASGVNAVALLVEE303 pKa = 4.53GTGAFLPAGSTLTLSFTASIPAGTLAGTVVRR334 pKa = 11.84NSAAATLDD342 pKa = 3.58GNGDD346 pKa = 3.62GDD348 pKa = 3.92GTDD351 pKa = 3.41PGEE354 pKa = 4.41TPVTPDD360 pKa = 2.61TTTTVGTVVGAAVGPAAFPQGGASGSYY387 pKa = 7.27TLRR390 pKa = 11.84GTAIDD395 pKa = 3.95RR396 pKa = 11.84SGDD399 pKa = 3.59TQTTTTPVTAGGNVTFRR416 pKa = 11.84QTVLNTGNASNDD428 pKa = 2.94FTLAVAGAPSGWTCTVQSISSSDD451 pKa = 3.52TLGTLTNPVTLSAGQSLDD469 pKa = 3.56FAVDD473 pKa = 3.27CALPYY478 pKa = 10.39GAAGTTNVVLTVSATPAGGSADD500 pKa = 3.66TTTSTVAQVSAAGAPLLGNGDD521 pKa = 4.83LNPATAPSQAGVTVATNPGTTASFPLEE548 pKa = 4.07LQNNGPVAEE557 pKa = 4.61TYY559 pKa = 9.57TLSSTLTGTQLYY571 pKa = 10.86ADD573 pKa = 4.47TNCDD577 pKa = 3.31GTPDD581 pKa = 3.49GAAITLTPSVAPGATLCLVAQQPVAAGTAAGEE613 pKa = 4.43TPVTFTATSTVTPSRR628 pKa = 11.84TTSVTDD634 pKa = 3.3TLRR637 pKa = 11.84VNAVVSGTFGPDD649 pKa = 3.14GAQSTIPGGSVTYY662 pKa = 10.85SHH664 pKa = 7.07TLTNTSNGAADD675 pKa = 3.87FTVLPFTSAQGFVYY689 pKa = 10.22SYY691 pKa = 9.17ATSSGGPFASTLSGTLAPGASTPLFVRR718 pKa = 11.84VNVPASTAGTPAEE731 pKa = 3.95AAAISVTLEE740 pKa = 3.84AQAAPQPAATLQVTDD755 pKa = 3.51TTTVQNVLASVVKK768 pKa = 9.93SVRR771 pKa = 11.84LCADD775 pKa = 3.68VSCTVTSAITGGQVSPGDD793 pKa = 3.54VLQYY797 pKa = 10.8TLTVQNSGSSPLNGAVLIDD816 pKa = 3.7TVPANTTFRR825 pKa = 11.84ALGGNNAALLYY836 pKa = 10.49SVDD839 pKa = 4.91GGGSWTATPPGALPTGDD856 pKa = 3.76FQAGLDD862 pKa = 3.84TNGDD866 pKa = 3.86NVVDD870 pKa = 4.06TADD873 pKa = 3.14ILAPGTSFEE882 pKa = 4.37VTFTVQVNN890 pKa = 3.38
MM1 pKa = 7.27SRR3 pKa = 11.84LSVRR7 pKa = 11.84LLCTVVGLSLLSSALAVGTPVNTSISNTATLNYY40 pKa = 9.69RR41 pKa = 11.84DD42 pKa = 4.47ALNAPRR48 pKa = 11.84TQTSNPVTVTVRR60 pKa = 11.84QVYY63 pKa = 10.07ALSLTPDD70 pKa = 3.46AAEE73 pKa = 3.88NAIPLSRR80 pKa = 11.84QFSAQAGASRR90 pKa = 11.84LIGYY94 pKa = 7.4EE95 pKa = 3.93LVNAGNGPDD104 pKa = 3.71TLTLGVTQSATDD116 pKa = 3.73GFDD119 pKa = 2.85VTTRR123 pKa = 11.84LLPDD127 pKa = 4.29ANCDD131 pKa = 3.46GTVDD135 pKa = 3.66SAVPLGAAQTLAADD149 pKa = 3.95EE150 pKa = 4.36RR151 pKa = 11.84LCVYY155 pKa = 10.53VEE157 pKa = 3.97ATVPAGAPTGAAARR171 pKa = 11.84LTLTAASQGNPAVTDD186 pKa = 3.68TEE188 pKa = 4.79NYY190 pKa = 9.9AQVTVSAAGQLDD202 pKa = 3.38ITKK205 pKa = 8.65TVSPTGSALPGAPLTYY221 pKa = 9.1TVSGSVPAGNPVGAVSGVLTVDD243 pKa = 3.93GVPRR247 pKa = 11.84SGVLVRR253 pKa = 11.84DD254 pKa = 3.59VLTTLEE260 pKa = 4.37FGAVTAASASSGAATALYY278 pKa = 8.97STDD281 pKa = 5.22GGATWTAAVPASGVNAVALLVEE303 pKa = 4.53GTGAFLPAGSTLTLSFTASIPAGTLAGTVVRR334 pKa = 11.84NSAAATLDD342 pKa = 3.58GNGDD346 pKa = 3.62GDD348 pKa = 3.92GTDD351 pKa = 3.41PGEE354 pKa = 4.41TPVTPDD360 pKa = 2.61TTTTVGTVVGAAVGPAAFPQGGASGSYY387 pKa = 7.27TLRR390 pKa = 11.84GTAIDD395 pKa = 3.95RR396 pKa = 11.84SGDD399 pKa = 3.59TQTTTTPVTAGGNVTFRR416 pKa = 11.84QTVLNTGNASNDD428 pKa = 2.94FTLAVAGAPSGWTCTVQSISSSDD451 pKa = 3.52TLGTLTNPVTLSAGQSLDD469 pKa = 3.56FAVDD473 pKa = 3.27CALPYY478 pKa = 10.39GAAGTTNVVLTVSATPAGGSADD500 pKa = 3.66TTTSTVAQVSAAGAPLLGNGDD521 pKa = 4.83LNPATAPSQAGVTVATNPGTTASFPLEE548 pKa = 4.07LQNNGPVAEE557 pKa = 4.61TYY559 pKa = 9.57TLSSTLTGTQLYY571 pKa = 10.86ADD573 pKa = 4.47TNCDD577 pKa = 3.31GTPDD581 pKa = 3.49GAAITLTPSVAPGATLCLVAQQPVAAGTAAGEE613 pKa = 4.43TPVTFTATSTVTPSRR628 pKa = 11.84TTSVTDD634 pKa = 3.3TLRR637 pKa = 11.84VNAVVSGTFGPDD649 pKa = 3.14GAQSTIPGGSVTYY662 pKa = 10.85SHH664 pKa = 7.07TLTNTSNGAADD675 pKa = 3.87FTVLPFTSAQGFVYY689 pKa = 10.22SYY691 pKa = 9.17ATSSGGPFASTLSGTLAPGASTPLFVRR718 pKa = 11.84VNVPASTAGTPAEE731 pKa = 3.95AAAISVTLEE740 pKa = 3.84AQAAPQPAATLQVTDD755 pKa = 3.51TTTVQNVLASVVKK768 pKa = 9.93SVRR771 pKa = 11.84LCADD775 pKa = 3.68VSCTVTSAITGGQVSPGDD793 pKa = 3.54VLQYY797 pKa = 10.8TLTVQNSGSSPLNGAVLIDD816 pKa = 3.7TVPANTTFRR825 pKa = 11.84ALGGNNAALLYY836 pKa = 10.49SVDD839 pKa = 4.91GGGSWTATPPGALPTGDD856 pKa = 3.76FQAGLDD862 pKa = 3.84TNGDD866 pKa = 3.86NVVDD870 pKa = 4.06TADD873 pKa = 3.14ILAPGTSFEE882 pKa = 4.37VTFTVQVNN890 pKa = 3.38
Molecular weight: 87.35 kDa
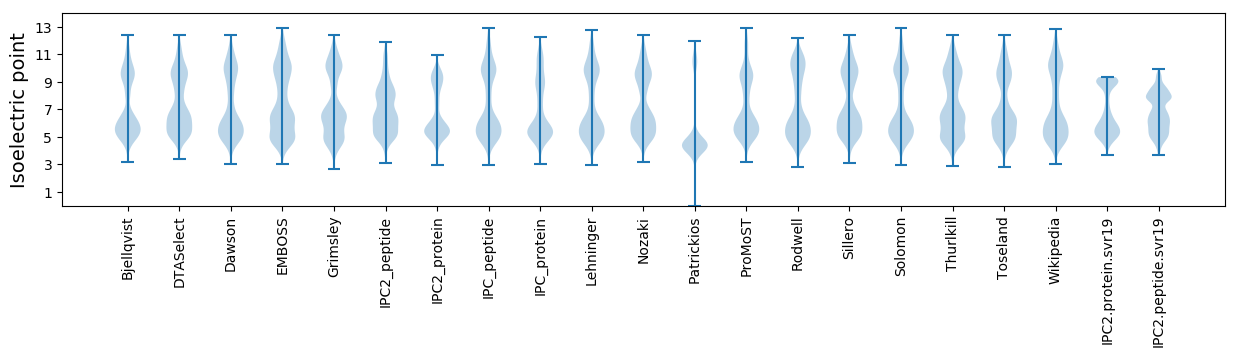
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A7C9HRY8|A0A7C9HRY8_9DEIO GIY-YIG nuclease family protein OS=Deinococcus sp. HMF7620 OX=2682977 GN=GO986_11375 PE=4 SV=1
MM1 pKa = 6.93NQRR4 pKa = 11.84NGALLAVLAMASVAAASPLTGGAQTLVMRR33 pKa = 11.84LARR36 pKa = 11.84AGDD39 pKa = 3.5VSMTGVRR46 pKa = 11.84WTEE49 pKa = 3.88DD50 pKa = 2.75GTTKK54 pKa = 10.35VVSRR58 pKa = 11.84AFNAQKK64 pKa = 10.9YY65 pKa = 9.52CLTEE69 pKa = 3.59PTLNLLQRR77 pKa = 11.84QPTNVGVQAMVFLEE91 pKa = 4.39FRR93 pKa = 11.84CVNRR97 pKa = 11.84IQGKK101 pKa = 9.67ALTPEE106 pKa = 4.0LALAFLRR113 pKa = 11.84SVGYY117 pKa = 8.02TVSGSNLLTNLLNNPPAANALATASQTAVTPMTPRR152 pKa = 11.84NN153 pKa = 3.49
MM1 pKa = 6.93NQRR4 pKa = 11.84NGALLAVLAMASVAAASPLTGGAQTLVMRR33 pKa = 11.84LARR36 pKa = 11.84AGDD39 pKa = 3.5VSMTGVRR46 pKa = 11.84WTEE49 pKa = 3.88DD50 pKa = 2.75GTTKK54 pKa = 10.35VVSRR58 pKa = 11.84AFNAQKK64 pKa = 10.9YY65 pKa = 9.52CLTEE69 pKa = 3.59PTLNLLQRR77 pKa = 11.84QPTNVGVQAMVFLEE91 pKa = 4.39FRR93 pKa = 11.84CVNRR97 pKa = 11.84IQGKK101 pKa = 9.67ALTPEE106 pKa = 4.0LALAFLRR113 pKa = 11.84SVGYY117 pKa = 8.02TVSGSNLLTNLLNNPPAANALATASQTAVTPMTPRR152 pKa = 11.84NN153 pKa = 3.49
Molecular weight: 16.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1325002 |
25 |
3293 |
304.8 |
32.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.047 ± 0.065 | 0.614 ± 0.011 |
5.051 ± 0.027 | 5.26 ± 0.036 |
2.976 ± 0.022 | 8.971 ± 0.045 |
2.195 ± 0.024 | 3.103 ± 0.033 |
2.354 ± 0.036 | 11.992 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.733 ± 0.018 | 2.343 ± 0.028 |
5.976 ± 0.042 | 4.326 ± 0.028 |
7.255 ± 0.037 | 4.967 ± 0.03 |
6.362 ± 0.035 | 7.708 ± 0.032 |
1.474 ± 0.019 | 2.293 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
