
Human papillomavirus KC5
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; unclassified Gammapapillomavirus
Average proteome isoelectric point is 6.15
Get precalculated fractions of proteins
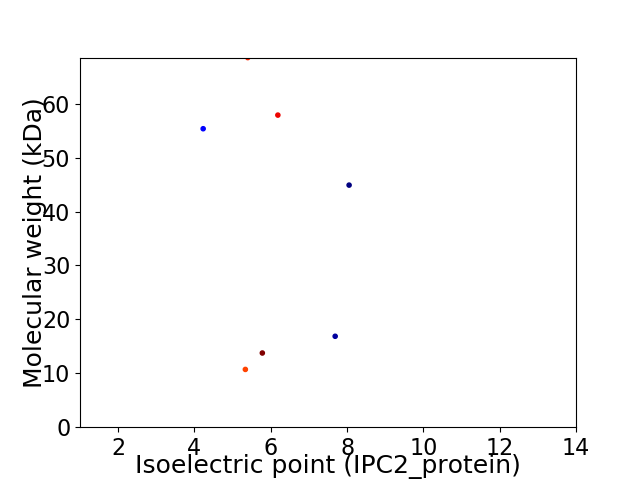
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|K4MNJ7|K4MNJ7_9PAPI Protein E7 OS=Human papillomavirus KC5 OX=1647924 GN=E7 PE=3 SV=1
MM1 pKa = 7.4SKK3 pKa = 9.74IRR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 9.71RR8 pKa = 11.84ASPEE12 pKa = 3.71DD13 pKa = 4.09LYY15 pKa = 11.12KK16 pKa = 10.67QCVLGADD23 pKa = 4.1CMPDD27 pKa = 3.06VKK29 pKa = 10.84NKK31 pKa = 10.45YY32 pKa = 7.38EE33 pKa = 4.06QNTLADD39 pKa = 3.34ILLKK43 pKa = 11.05VFGSLVYY50 pKa = 10.32FGGLGIGSGRR60 pKa = 11.84GTGGRR65 pKa = 11.84LGYY68 pKa = 10.2RR69 pKa = 11.84PLGEE73 pKa = 4.64AGGSRR78 pKa = 11.84SGRR81 pKa = 11.84PITSTPVRR89 pKa = 11.84PSVTVDD95 pKa = 2.94VLGPSDD101 pKa = 4.04IVPVDD106 pKa = 3.47AEE108 pKa = 4.4TPAIVPLAEE117 pKa = 4.31GTPDD121 pKa = 3.9LDD123 pKa = 4.73FVAPDD128 pKa = 3.82AGPGVGVEE136 pKa = 4.06EE137 pKa = 4.78LEE139 pKa = 4.95LYY141 pKa = 9.93TITDD145 pKa = 3.51STATTGGVTQSTTVTTNEE163 pKa = 3.65TSFAVLDD170 pKa = 3.83VNTIGEE176 pKa = 4.15KK177 pKa = 10.07PIQVFYY183 pKa = 11.34DD184 pKa = 3.98PAAPDD189 pKa = 3.72TQILNVFTASPANTSDD205 pKa = 2.8INIFVDD211 pKa = 4.49SSITGQTIGEE221 pKa = 4.33YY222 pKa = 10.58EE223 pKa = 4.57SIPLEE228 pKa = 3.95RR229 pKa = 11.84LDD231 pKa = 3.6YY232 pKa = 11.38SEE234 pKa = 5.87FEE236 pKa = 4.08IEE238 pKa = 4.31EE239 pKa = 4.7PPTTSTPSQRR249 pKa = 11.84LDD251 pKa = 3.24RR252 pKa = 11.84TINRR256 pKa = 11.84ARR258 pKa = 11.84QLYY261 pKa = 9.36NRR263 pKa = 11.84FVRR266 pKa = 11.84QVPVSSAEE274 pKa = 4.0FVTQPSRR281 pKa = 11.84LVQFEE286 pKa = 4.51FEE288 pKa = 4.04NAAYY292 pKa = 10.13DD293 pKa = 3.65ADD295 pKa = 3.7VSIEE299 pKa = 3.91FEE301 pKa = 3.85RR302 pKa = 11.84DD303 pKa = 3.08LAEE306 pKa = 4.29VAAAPDD312 pKa = 3.35NAFRR316 pKa = 11.84DD317 pKa = 4.01VTRR320 pKa = 11.84LHH322 pKa = 6.8RR323 pKa = 11.84PILTEE328 pKa = 3.61TSEE331 pKa = 4.62GIVRR335 pKa = 11.84VSRR338 pKa = 11.84LAEE341 pKa = 4.02TGTITTRR348 pKa = 11.84SGTTIGQPIHH358 pKa = 6.76FYY360 pKa = 11.15YY361 pKa = 10.51DD362 pKa = 2.96ISEE365 pKa = 4.26IPPIEE370 pKa = 5.14DD371 pKa = 4.3IEE373 pKa = 4.37LQTIAGQSNTATATIVDD390 pKa = 5.38DD391 pKa = 3.98ILATTVIDD399 pKa = 5.0PINATDD405 pKa = 4.09VIHH408 pKa = 7.03NDD410 pKa = 4.23DD411 pKa = 3.86ILEE414 pKa = 4.34DD415 pKa = 3.98EE416 pKa = 4.51YY417 pKa = 11.96AEE419 pKa = 4.42SFNNAQLVIQSTIEE433 pKa = 4.06EE434 pKa = 4.48GDD436 pKa = 3.93SLIMPTFAPEE446 pKa = 3.54ASIKK450 pKa = 10.8VFVNDD455 pKa = 3.9LGGGLFVTYY464 pKa = 9.52PSGSDD469 pKa = 3.05SSNIIILPNDD479 pKa = 3.4IPIGPPVIIDD489 pKa = 3.76AFDD492 pKa = 4.68DD493 pKa = 4.76YY494 pKa = 11.11ILHH497 pKa = 6.98PAVIPKK503 pKa = 9.21KK504 pKa = 9.79RR505 pKa = 11.84RR506 pKa = 11.84RR507 pKa = 11.84IEE509 pKa = 3.88LFF511 pKa = 3.27
MM1 pKa = 7.4SKK3 pKa = 9.74IRR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 9.71RR8 pKa = 11.84ASPEE12 pKa = 3.71DD13 pKa = 4.09LYY15 pKa = 11.12KK16 pKa = 10.67QCVLGADD23 pKa = 4.1CMPDD27 pKa = 3.06VKK29 pKa = 10.84NKK31 pKa = 10.45YY32 pKa = 7.38EE33 pKa = 4.06QNTLADD39 pKa = 3.34ILLKK43 pKa = 11.05VFGSLVYY50 pKa = 10.32FGGLGIGSGRR60 pKa = 11.84GTGGRR65 pKa = 11.84LGYY68 pKa = 10.2RR69 pKa = 11.84PLGEE73 pKa = 4.64AGGSRR78 pKa = 11.84SGRR81 pKa = 11.84PITSTPVRR89 pKa = 11.84PSVTVDD95 pKa = 2.94VLGPSDD101 pKa = 4.04IVPVDD106 pKa = 3.47AEE108 pKa = 4.4TPAIVPLAEE117 pKa = 4.31GTPDD121 pKa = 3.9LDD123 pKa = 4.73FVAPDD128 pKa = 3.82AGPGVGVEE136 pKa = 4.06EE137 pKa = 4.78LEE139 pKa = 4.95LYY141 pKa = 9.93TITDD145 pKa = 3.51STATTGGVTQSTTVTTNEE163 pKa = 3.65TSFAVLDD170 pKa = 3.83VNTIGEE176 pKa = 4.15KK177 pKa = 10.07PIQVFYY183 pKa = 11.34DD184 pKa = 3.98PAAPDD189 pKa = 3.72TQILNVFTASPANTSDD205 pKa = 2.8INIFVDD211 pKa = 4.49SSITGQTIGEE221 pKa = 4.33YY222 pKa = 10.58EE223 pKa = 4.57SIPLEE228 pKa = 3.95RR229 pKa = 11.84LDD231 pKa = 3.6YY232 pKa = 11.38SEE234 pKa = 5.87FEE236 pKa = 4.08IEE238 pKa = 4.31EE239 pKa = 4.7PPTTSTPSQRR249 pKa = 11.84LDD251 pKa = 3.24RR252 pKa = 11.84TINRR256 pKa = 11.84ARR258 pKa = 11.84QLYY261 pKa = 9.36NRR263 pKa = 11.84FVRR266 pKa = 11.84QVPVSSAEE274 pKa = 4.0FVTQPSRR281 pKa = 11.84LVQFEE286 pKa = 4.51FEE288 pKa = 4.04NAAYY292 pKa = 10.13DD293 pKa = 3.65ADD295 pKa = 3.7VSIEE299 pKa = 3.91FEE301 pKa = 3.85RR302 pKa = 11.84DD303 pKa = 3.08LAEE306 pKa = 4.29VAAAPDD312 pKa = 3.35NAFRR316 pKa = 11.84DD317 pKa = 4.01VTRR320 pKa = 11.84LHH322 pKa = 6.8RR323 pKa = 11.84PILTEE328 pKa = 3.61TSEE331 pKa = 4.62GIVRR335 pKa = 11.84VSRR338 pKa = 11.84LAEE341 pKa = 4.02TGTITTRR348 pKa = 11.84SGTTIGQPIHH358 pKa = 6.76FYY360 pKa = 11.15YY361 pKa = 10.51DD362 pKa = 2.96ISEE365 pKa = 4.26IPPIEE370 pKa = 5.14DD371 pKa = 4.3IEE373 pKa = 4.37LQTIAGQSNTATATIVDD390 pKa = 5.38DD391 pKa = 3.98ILATTVIDD399 pKa = 5.0PINATDD405 pKa = 4.09VIHH408 pKa = 7.03NDD410 pKa = 4.23DD411 pKa = 3.86ILEE414 pKa = 4.34DD415 pKa = 3.98EE416 pKa = 4.51YY417 pKa = 11.96AEE419 pKa = 4.42SFNNAQLVIQSTIEE433 pKa = 4.06EE434 pKa = 4.48GDD436 pKa = 3.93SLIMPTFAPEE446 pKa = 3.54ASIKK450 pKa = 10.8VFVNDD455 pKa = 3.9LGGGLFVTYY464 pKa = 9.52PSGSDD469 pKa = 3.05SSNIIILPNDD479 pKa = 3.4IPIGPPVIIDD489 pKa = 3.76AFDD492 pKa = 4.68DD493 pKa = 4.76YY494 pKa = 11.11ILHH497 pKa = 6.98PAVIPKK503 pKa = 9.21KK504 pKa = 9.79RR505 pKa = 11.84RR506 pKa = 11.84RR507 pKa = 11.84IEE509 pKa = 3.88LFF511 pKa = 3.27
Molecular weight: 55.36 kDa
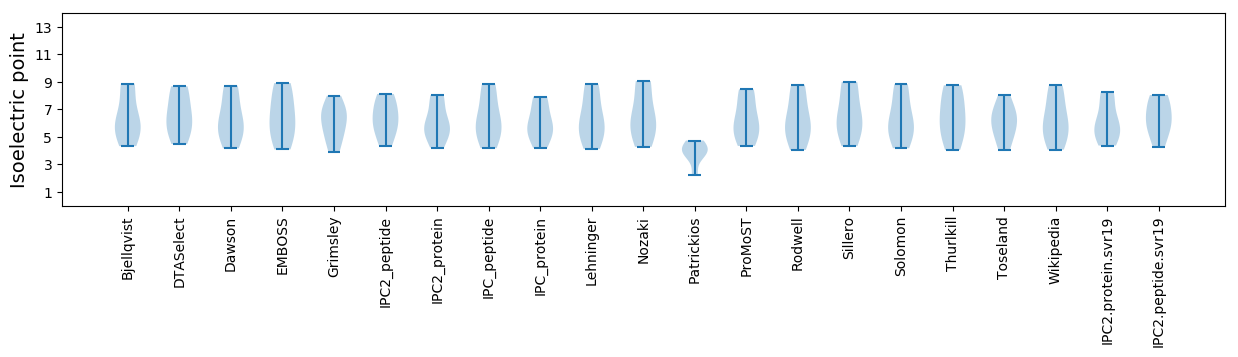
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K4MKX9|K4MKX9_9PAPI Major capsid protein L1 OS=Human papillomavirus KC5 OX=1647924 GN=L1 PE=3 SV=1
MM1 pKa = 7.42EE2 pKa = 5.25SKK4 pKa = 10.28PRR6 pKa = 11.84NLEE9 pKa = 4.2EE10 pKa = 4.05YY11 pKa = 8.89CKK13 pKa = 10.46HH14 pKa = 5.88YY15 pKa = 10.77GISFFNLQLRR25 pKa = 11.84CVFCHH30 pKa = 5.83HH31 pKa = 7.4WIDD34 pKa = 4.44SIQLASFHH42 pKa = 6.62CKK44 pKa = 9.18QLALIWKK51 pKa = 9.59NNVCFAACTQCLKK64 pKa = 10.9LSAAHH69 pKa = 5.42EE70 pKa = 4.12RR71 pKa = 11.84EE72 pKa = 3.79RR73 pKa = 11.84FYY75 pKa = 11.27QCNVRR80 pKa = 11.84SDD82 pKa = 3.89LLADD86 pKa = 4.8FLHH89 pKa = 6.82KK90 pKa = 10.27PLNDD94 pKa = 3.26VVIRR98 pKa = 11.84CLYY101 pKa = 10.12CLRR104 pKa = 11.84KK105 pKa = 9.76LDD107 pKa = 4.62FIEE110 pKa = 5.05KK111 pKa = 10.54LEE113 pKa = 4.01HH114 pKa = 7.14CYY116 pKa = 11.14NKK118 pKa = 10.66DD119 pKa = 3.17CFHH122 pKa = 7.46LVRR125 pKa = 11.84CYY127 pKa = 9.8WRR129 pKa = 11.84GKK131 pKa = 9.78CRR133 pKa = 11.84NCKK136 pKa = 9.87YY137 pKa = 11.0NEE139 pKa = 4.04GG140 pKa = 3.74
MM1 pKa = 7.42EE2 pKa = 5.25SKK4 pKa = 10.28PRR6 pKa = 11.84NLEE9 pKa = 4.2EE10 pKa = 4.05YY11 pKa = 8.89CKK13 pKa = 10.46HH14 pKa = 5.88YY15 pKa = 10.77GISFFNLQLRR25 pKa = 11.84CVFCHH30 pKa = 5.83HH31 pKa = 7.4WIDD34 pKa = 4.44SIQLASFHH42 pKa = 6.62CKK44 pKa = 9.18QLALIWKK51 pKa = 9.59NNVCFAACTQCLKK64 pKa = 10.9LSAAHH69 pKa = 5.42EE70 pKa = 4.12RR71 pKa = 11.84EE72 pKa = 3.79RR73 pKa = 11.84FYY75 pKa = 11.27QCNVRR80 pKa = 11.84SDD82 pKa = 3.89LLADD86 pKa = 4.8FLHH89 pKa = 6.82KK90 pKa = 10.27PLNDD94 pKa = 3.26VVIRR98 pKa = 11.84CLYY101 pKa = 10.12CLRR104 pKa = 11.84KK105 pKa = 9.76LDD107 pKa = 4.62FIEE110 pKa = 5.05KK111 pKa = 10.54LEE113 pKa = 4.01HH114 pKa = 7.14CYY116 pKa = 11.14NKK118 pKa = 10.66DD119 pKa = 3.17CFHH122 pKa = 7.46LVRR125 pKa = 11.84CYY127 pKa = 9.8WRR129 pKa = 11.84GKK131 pKa = 9.78CRR133 pKa = 11.84NCKK136 pKa = 9.87YY137 pKa = 11.0NEE139 pKa = 4.04GG140 pKa = 3.74
Molecular weight: 16.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2364 |
93 |
600 |
337.7 |
38.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.795 ± 0.372 | 2.411 ± 0.945 |
5.964 ± 0.487 | 7.064 ± 0.515 |
5.161 ± 0.442 | 5.203 ± 0.766 |
2.2 ± 0.428 | 5.668 ± 0.865 |
5.626 ± 0.827 | 8.756 ± 0.736 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.481 ± 0.429 | 4.822 ± 0.541 |
6.303 ± 1.025 | 3.723 ± 0.263 |
5.541 ± 0.602 | 7.149 ± 0.828 |
6.514 ± 0.966 | 5.922 ± 0.492 |
1.311 ± 0.289 | 3.384 ± 0.328 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
