
Deinococcus sp. K2S05-167
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Deinococcus-Thermus; Deinococci; Deinococcales; Deinococcaceae; Deinococcus; unclassified Deinococcus
Average proteome isoelectric point is 6.82
Get precalculated fractions of proteins
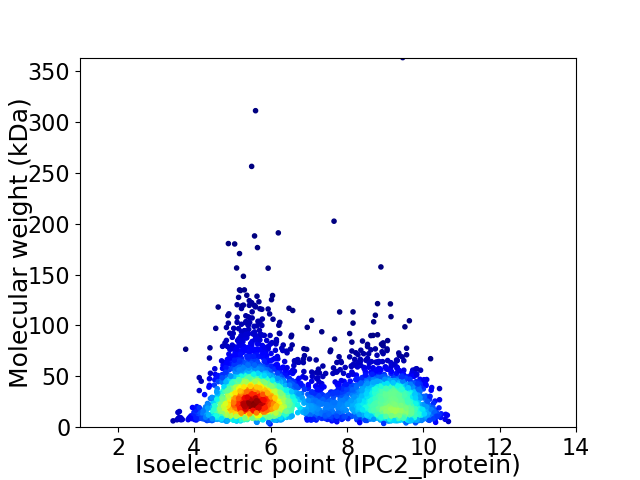
Virtual 2D-PAGE plot for 4078 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A418V806|A0A418V806_9DEIO Dihydrolipoamide acetyltransferase component of pyruvate dehydrogenase complex OS=Deinococcus sp. K2S05-167 OX=2320857 GN=D3875_12345 PE=3 SV=1
MM1 pKa = 7.61KK2 pKa = 10.62LNPKK6 pKa = 9.83LLALMTALAAGAASAAPPAPNTTSTGAGVTISNTATATFTDD47 pKa = 4.26PATNAAATPVQSNTVDD63 pKa = 3.61TVVLPQPGFDD73 pKa = 2.96IVYY76 pKa = 10.75ADD78 pKa = 3.77GTNDD82 pKa = 3.55GNTLGNTTKK91 pKa = 10.18TVTNATPGQSNSTDD105 pKa = 3.33YY106 pKa = 11.49YY107 pKa = 10.93VVNNGNTPLTVQVTANTSGSASGATVTYY135 pKa = 10.83LDD137 pKa = 3.81ASGNALPSSTTGGVTTYY154 pKa = 10.32TVTLPAGAAGMVKK167 pKa = 8.51ITQVITLPGNAPANTTYY184 pKa = 10.74GASPEE189 pKa = 4.22GTVGGTGTGTGQNGIPTGSTLYY211 pKa = 10.58EE212 pKa = 4.19NQTVTTTNGTTTVNGTPAQGADD234 pKa = 3.5LQFVKK239 pKa = 9.83LTTFKK244 pKa = 10.41PSLSATPNDD253 pKa = 4.21PNTPNPTAPDD263 pKa = 3.65GTTPATPPTLGTVQVPTVPAGTANTPNPTNPAQGYY298 pKa = 9.93LSTPSNPGDD307 pKa = 3.69PTSGGTPIVPDD318 pKa = 3.56LTGNTQIAYY327 pKa = 8.61PKK329 pKa = 10.25ADD331 pKa = 4.1PDD333 pKa = 4.1NNPTTIPAAGQTNDD347 pKa = 3.45QPGSADD353 pKa = 3.2IINFTNEE360 pKa = 3.51VKK362 pKa = 10.84NAGTVSDD369 pKa = 4.55KK370 pKa = 10.97VQLYY374 pKa = 8.68PAGPDD379 pKa = 3.27GKK381 pKa = 10.41LLAGTTFDD389 pKa = 4.04ASTGTFTLPGGIQVVFLGPNTNAAIPVASGATYY422 pKa = 7.62PTLTVPAGGSAVFRR436 pKa = 11.84TQVITTDD443 pKa = 3.48ANDD446 pKa = 3.72NLLTNAINIVVGADD460 pKa = 3.4SLNDD464 pKa = 3.46ADD466 pKa = 5.19IIADD470 pKa = 3.61ATTTDD475 pKa = 3.73VILPAAAQFGDD486 pKa = 4.1SNGTSPLGAVPTPAPVQQVTPSSNTSPSTDD516 pKa = 3.46PNLSSDD522 pKa = 3.66NVAVFPMDD530 pKa = 3.47VANLGQYY537 pKa = 8.27TDD539 pKa = 4.44SYY541 pKa = 9.77TLSATVAGLPAGATISYY558 pKa = 9.57VNSSGVALPTNGAGKK573 pKa = 10.05FVTPVVAAGQEE584 pKa = 3.94IVVYY588 pKa = 10.45AVVTVPTGTAAGNYY602 pKa = 6.83TVKK605 pKa = 10.36QQAVGNYY612 pKa = 7.15STITMTDD619 pKa = 3.25LNDD622 pKa = 3.72VIKK625 pKa = 10.75VGAVGDD631 pKa = 3.63VALAKK636 pKa = 10.17FVQDD640 pKa = 3.84GKK642 pKa = 9.6TSAGSNSYY650 pKa = 11.1AGINNPQNYY659 pKa = 7.5TVNNTSALPGTNIVYY674 pKa = 9.85QIIGKK679 pKa = 9.14NNYY682 pKa = 8.8NSPVANFALNDD693 pKa = 3.87SVPPNTTFQGAVLTINGTAVSRR715 pKa = 11.84VIYY718 pKa = 10.28KK719 pKa = 10.34VGAGTWSTTAPTVGTPAGTSIAVAADD745 pKa = 3.61TDD747 pKa = 3.95SDD749 pKa = 4.6NIPDD753 pKa = 4.46ALPAGSSMEE762 pKa = 3.99LVFTVKK768 pKa = 10.4VNN770 pKa = 3.28
MM1 pKa = 7.61KK2 pKa = 10.62LNPKK6 pKa = 9.83LLALMTALAAGAASAAPPAPNTTSTGAGVTISNTATATFTDD47 pKa = 4.26PATNAAATPVQSNTVDD63 pKa = 3.61TVVLPQPGFDD73 pKa = 2.96IVYY76 pKa = 10.75ADD78 pKa = 3.77GTNDD82 pKa = 3.55GNTLGNTTKK91 pKa = 10.18TVTNATPGQSNSTDD105 pKa = 3.33YY106 pKa = 11.49YY107 pKa = 10.93VVNNGNTPLTVQVTANTSGSASGATVTYY135 pKa = 10.83LDD137 pKa = 3.81ASGNALPSSTTGGVTTYY154 pKa = 10.32TVTLPAGAAGMVKK167 pKa = 8.51ITQVITLPGNAPANTTYY184 pKa = 10.74GASPEE189 pKa = 4.22GTVGGTGTGTGQNGIPTGSTLYY211 pKa = 10.58EE212 pKa = 4.19NQTVTTTNGTTTVNGTPAQGADD234 pKa = 3.5LQFVKK239 pKa = 9.83LTTFKK244 pKa = 10.41PSLSATPNDD253 pKa = 4.21PNTPNPTAPDD263 pKa = 3.65GTTPATPPTLGTVQVPTVPAGTANTPNPTNPAQGYY298 pKa = 9.93LSTPSNPGDD307 pKa = 3.69PTSGGTPIVPDD318 pKa = 3.56LTGNTQIAYY327 pKa = 8.61PKK329 pKa = 10.25ADD331 pKa = 4.1PDD333 pKa = 4.1NNPTTIPAAGQTNDD347 pKa = 3.45QPGSADD353 pKa = 3.2IINFTNEE360 pKa = 3.51VKK362 pKa = 10.84NAGTVSDD369 pKa = 4.55KK370 pKa = 10.97VQLYY374 pKa = 8.68PAGPDD379 pKa = 3.27GKK381 pKa = 10.41LLAGTTFDD389 pKa = 4.04ASTGTFTLPGGIQVVFLGPNTNAAIPVASGATYY422 pKa = 7.62PTLTVPAGGSAVFRR436 pKa = 11.84TQVITTDD443 pKa = 3.48ANDD446 pKa = 3.72NLLTNAINIVVGADD460 pKa = 3.4SLNDD464 pKa = 3.46ADD466 pKa = 5.19IIADD470 pKa = 3.61ATTTDD475 pKa = 3.73VILPAAAQFGDD486 pKa = 4.1SNGTSPLGAVPTPAPVQQVTPSSNTSPSTDD516 pKa = 3.46PNLSSDD522 pKa = 3.66NVAVFPMDD530 pKa = 3.47VANLGQYY537 pKa = 8.27TDD539 pKa = 4.44SYY541 pKa = 9.77TLSATVAGLPAGATISYY558 pKa = 9.57VNSSGVALPTNGAGKK573 pKa = 10.05FVTPVVAAGQEE584 pKa = 3.94IVVYY588 pKa = 10.45AVVTVPTGTAAGNYY602 pKa = 6.83TVKK605 pKa = 10.36QQAVGNYY612 pKa = 7.15STITMTDD619 pKa = 3.25LNDD622 pKa = 3.72VIKK625 pKa = 10.75VGAVGDD631 pKa = 3.63VALAKK636 pKa = 10.17FVQDD640 pKa = 3.84GKK642 pKa = 9.6TSAGSNSYY650 pKa = 11.1AGINNPQNYY659 pKa = 7.5TVNNTSALPGTNIVYY674 pKa = 9.85QIIGKK679 pKa = 9.14NNYY682 pKa = 8.8NSPVANFALNDD693 pKa = 3.87SVPPNTTFQGAVLTINGTAVSRR715 pKa = 11.84VIYY718 pKa = 10.28KK719 pKa = 10.34VGAGTWSTTAPTVGTPAGTSIAVAADD745 pKa = 3.61TDD747 pKa = 3.95SDD749 pKa = 4.6NIPDD753 pKa = 4.46ALPAGSSMEE762 pKa = 3.99LVFTVKK768 pKa = 10.4VNN770 pKa = 3.28
Molecular weight: 76.62 kDa
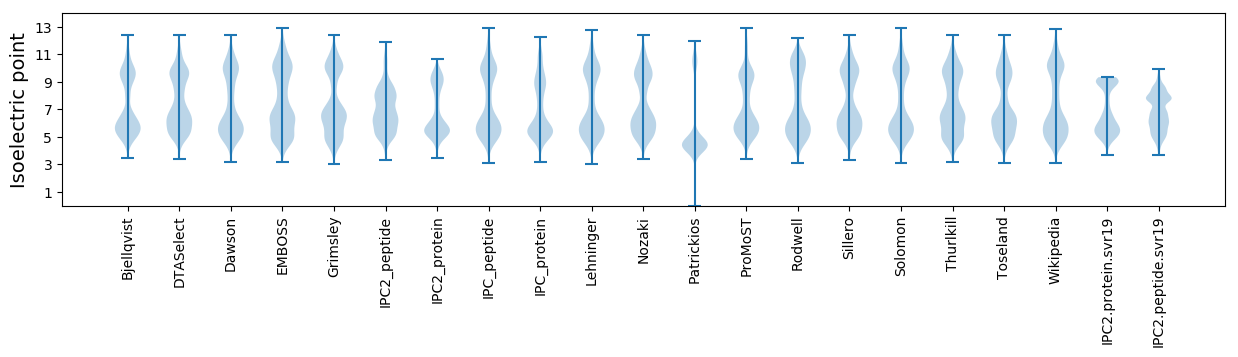
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A418V7P3|A0A418V7P3_9DEIO Uncharacterized protein OS=Deinococcus sp. K2S05-167 OX=2320857 GN=D3875_11680 PE=4 SV=1
MM1 pKa = 7.25NRR3 pKa = 11.84NFTPEE8 pKa = 3.85LSSVYY13 pKa = 9.74RR14 pKa = 11.84DD15 pKa = 3.35EE16 pKa = 4.56GRR18 pKa = 11.84QISVALRR25 pKa = 11.84PGNYY29 pKa = 8.48TFLVMARR36 pKa = 11.84DD37 pKa = 3.6ARR39 pKa = 11.84TGRR42 pKa = 11.84TLWKK46 pKa = 10.13FHH48 pKa = 5.65GQGTASVDD56 pKa = 3.05ARR58 pKa = 11.84LAGQGAVMVTVITTGAITRR77 pKa = 11.84SQALLLDD84 pKa = 3.72ARR86 pKa = 11.84TGSVRR91 pKa = 11.84RR92 pKa = 11.84KK93 pKa = 10.48GLFTLEE99 pKa = 3.99GVRR102 pKa = 11.84DD103 pKa = 4.18GKK105 pKa = 11.03ALFMQYY111 pKa = 11.08DD112 pKa = 3.93EE113 pKa = 5.56APPSAAFLADD123 pKa = 3.85PNVLLGEE130 pKa = 4.28VMQVRR135 pKa = 11.84TGRR138 pKa = 11.84TLTRR142 pKa = 11.84NLPIPTRR149 pKa = 11.84PGCGPLKK156 pKa = 9.06TLKK159 pKa = 10.63VGSNMQAFRR168 pKa = 11.84MNDD171 pKa = 3.29RR172 pKa = 11.84QQLVATRR179 pKa = 11.84QDD181 pKa = 2.96RR182 pKa = 11.84CGTFQATFDD191 pKa = 3.33WWKK194 pKa = 10.76VPLPAPKK201 pKa = 9.53IEE203 pKa = 4.38SSASS207 pKa = 3.19
MM1 pKa = 7.25NRR3 pKa = 11.84NFTPEE8 pKa = 3.85LSSVYY13 pKa = 9.74RR14 pKa = 11.84DD15 pKa = 3.35EE16 pKa = 4.56GRR18 pKa = 11.84QISVALRR25 pKa = 11.84PGNYY29 pKa = 8.48TFLVMARR36 pKa = 11.84DD37 pKa = 3.6ARR39 pKa = 11.84TGRR42 pKa = 11.84TLWKK46 pKa = 10.13FHH48 pKa = 5.65GQGTASVDD56 pKa = 3.05ARR58 pKa = 11.84LAGQGAVMVTVITTGAITRR77 pKa = 11.84SQALLLDD84 pKa = 3.72ARR86 pKa = 11.84TGSVRR91 pKa = 11.84RR92 pKa = 11.84KK93 pKa = 10.48GLFTLEE99 pKa = 3.99GVRR102 pKa = 11.84DD103 pKa = 4.18GKK105 pKa = 11.03ALFMQYY111 pKa = 11.08DD112 pKa = 3.93EE113 pKa = 5.56APPSAAFLADD123 pKa = 3.85PNVLLGEE130 pKa = 4.28VMQVRR135 pKa = 11.84TGRR138 pKa = 11.84TLTRR142 pKa = 11.84NLPIPTRR149 pKa = 11.84PGCGPLKK156 pKa = 9.06TLKK159 pKa = 10.63VGSNMQAFRR168 pKa = 11.84MNDD171 pKa = 3.29RR172 pKa = 11.84QQLVATRR179 pKa = 11.84QDD181 pKa = 2.96RR182 pKa = 11.84CGTFQATFDD191 pKa = 3.33WWKK194 pKa = 10.76VPLPAPKK201 pKa = 9.53IEE203 pKa = 4.38SSASS207 pKa = 3.19
Molecular weight: 22.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1205993 |
28 |
3540 |
295.7 |
32.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.435 ± 0.043 | 0.632 ± 0.012 |
5.034 ± 0.028 | 5.738 ± 0.038 |
3.281 ± 0.023 | 8.525 ± 0.043 |
2.326 ± 0.021 | 3.753 ± 0.029 |
3.217 ± 0.034 | 11.516 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.059 ± 0.019 | 2.785 ± 0.024 |
5.662 ± 0.032 | 4.454 ± 0.031 |
7.029 ± 0.039 | 5.157 ± 0.025 |
6.033 ± 0.036 | 7.56 ± 0.034 |
1.417 ± 0.017 | 2.386 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
