
Cucumber leaf spot virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Procedovirinae; Aureusvirus
Average proteome isoelectric point is 7.72
Get precalculated fractions of proteins
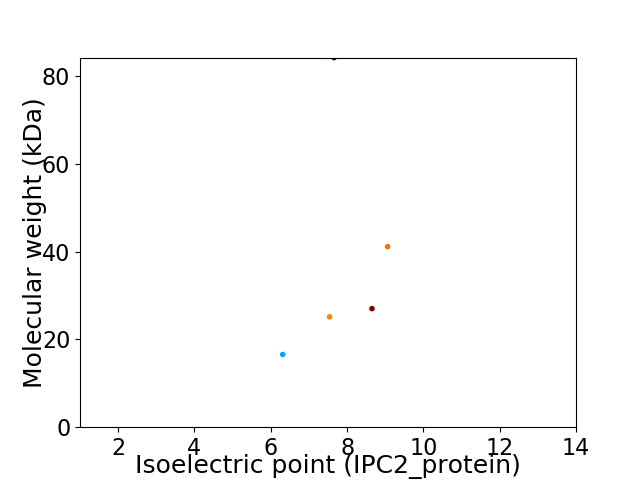
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A8CZ23|A8CZ23_9TOMB p17 OS=Cucumber leaf spot virus OX=165432 PE=4 SV=1
MM1 pKa = 7.86EE2 pKa = 4.39NTQGGVLCPNRR13 pKa = 11.84CKK15 pKa = 10.67VCSHH19 pKa = 5.91TTCIRR24 pKa = 11.84EE25 pKa = 3.96SSGQGGRR32 pKa = 11.84QASRR36 pKa = 11.84FTRR39 pKa = 11.84LITEE43 pKa = 3.8PRR45 pKa = 11.84IVSEE49 pKa = 3.89QGIQYY54 pKa = 7.55RR55 pKa = 11.84TWIADD60 pKa = 3.32RR61 pKa = 11.84GFPIALLSSSGGLSTSIRR79 pKa = 11.84GHH81 pKa = 5.29GVAVTVQGDD90 pKa = 4.09SKK92 pKa = 11.61SLLNFCRR99 pKa = 11.84VAYY102 pKa = 10.11DD103 pKa = 3.52VFNHH107 pKa = 6.02PVIQSEE113 pKa = 4.62VCYY116 pKa = 10.72GRR118 pKa = 11.84GPSTSDD124 pKa = 2.86EE125 pKa = 4.07VRR127 pKa = 11.84AEE129 pKa = 4.13FQDD132 pKa = 3.4EE133 pKa = 3.98AGIIQGRR140 pKa = 11.84LPQSEE145 pKa = 4.32TEE147 pKa = 4.39EE148 pKa = 4.1NNKK151 pKa = 9.94AA152 pKa = 3.31
MM1 pKa = 7.86EE2 pKa = 4.39NTQGGVLCPNRR13 pKa = 11.84CKK15 pKa = 10.67VCSHH19 pKa = 5.91TTCIRR24 pKa = 11.84EE25 pKa = 3.96SSGQGGRR32 pKa = 11.84QASRR36 pKa = 11.84FTRR39 pKa = 11.84LITEE43 pKa = 3.8PRR45 pKa = 11.84IVSEE49 pKa = 3.89QGIQYY54 pKa = 7.55RR55 pKa = 11.84TWIADD60 pKa = 3.32RR61 pKa = 11.84GFPIALLSSSGGLSTSIRR79 pKa = 11.84GHH81 pKa = 5.29GVAVTVQGDD90 pKa = 4.09SKK92 pKa = 11.61SLLNFCRR99 pKa = 11.84VAYY102 pKa = 10.11DD103 pKa = 3.52VFNHH107 pKa = 6.02PVIQSEE113 pKa = 4.62VCYY116 pKa = 10.72GRR118 pKa = 11.84GPSTSDD124 pKa = 2.86EE125 pKa = 4.07VRR127 pKa = 11.84AEE129 pKa = 4.13FQDD132 pKa = 3.4EE133 pKa = 3.98AGIIQGRR140 pKa = 11.84LPQSEE145 pKa = 4.32TEE147 pKa = 4.39EE148 pKa = 4.1NNKK151 pKa = 9.94AA152 pKa = 3.31
Molecular weight: 16.56 kDa
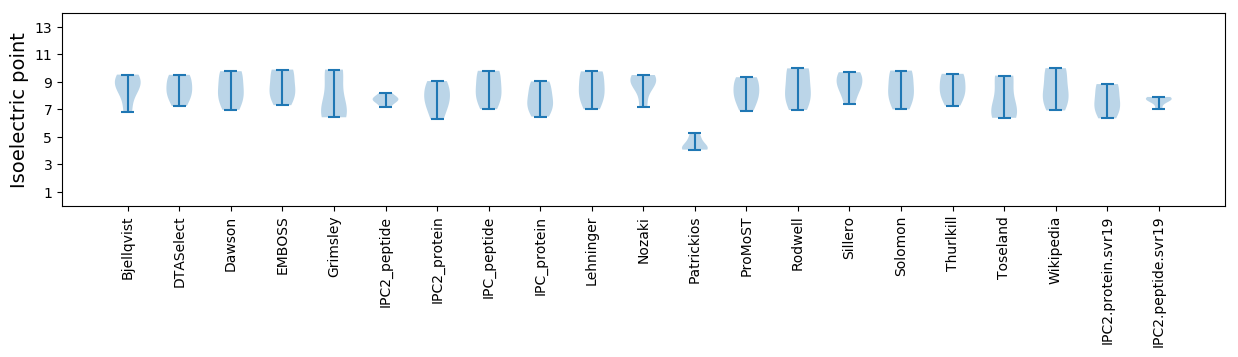
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A8CZ21|A8CZ21_9TOMB p27 OS=Cucumber leaf spot virus OX=165432 PE=3 SV=1
MM1 pKa = 7.66EE2 pKa = 5.1IARR5 pKa = 11.84TNKK8 pKa = 9.71NSVVKK13 pKa = 8.68YY14 pKa = 9.71VPAAVGAAYY23 pKa = 10.58QMGKK27 pKa = 10.27SIVPYY32 pKa = 10.54APTIVDD38 pKa = 3.15ALGNVVSRR46 pKa = 11.84ATGRR50 pKa = 11.84KK51 pKa = 8.67KK52 pKa = 10.16KK53 pKa = 10.74SKK55 pKa = 10.31GKK57 pKa = 9.45EE58 pKa = 3.73VQNQIVGGIGAIAAPVSITKK78 pKa = 10.12RR79 pKa = 11.84VRR81 pKa = 11.84GMRR84 pKa = 11.84PSFRR88 pKa = 11.84QTKK91 pKa = 8.35GKK93 pKa = 8.67VHH95 pKa = 5.99IVHH98 pKa = 6.85RR99 pKa = 11.84EE100 pKa = 3.74LVTSVINLVGNFRR113 pKa = 11.84VNNNVSAQIGQFRR126 pKa = 11.84INPSNSSLFTWLPTIASNFDD146 pKa = 3.38SYY148 pKa = 11.49RR149 pKa = 11.84FTSIRR154 pKa = 11.84FVYY157 pKa = 10.57VPLCATTEE165 pKa = 4.01TGRR168 pKa = 11.84VSLFWDD174 pKa = 4.23KK175 pKa = 11.29DD176 pKa = 3.79SQDD179 pKa = 3.91PLPVDD184 pKa = 4.13RR185 pKa = 11.84AALSSYY191 pKa = 9.48GHH193 pKa = 6.23SNEE196 pKa = 3.9GPPWAEE202 pKa = 3.7TTLNVPTDD210 pKa = 3.55GKK212 pKa = 10.76QRR214 pKa = 11.84FVTDD218 pKa = 3.13SNTTDD223 pKa = 3.11RR224 pKa = 11.84KK225 pKa = 10.55LVDD228 pKa = 3.99LGQFAFATYY237 pKa = 10.74AGGSNNQIGDD247 pKa = 3.43IYY249 pKa = 11.18VEE251 pKa = 4.09YY252 pKa = 10.57GVEE255 pKa = 4.13FSEE258 pKa = 4.33AQPAGGLTQYY268 pKa = 8.43ITKK271 pKa = 10.37SVGATASTTGPSYY284 pKa = 11.42VVDD287 pKa = 3.87ANINVNATTANVEE300 pKa = 4.27FFSPGTFLITAVVYY314 pKa = 10.53GSTIASPSMAGGNGTLIGDD333 pKa = 4.35LPVVGGSNASIWTCVFSTTGVSTSVPTFTQAGTGLTRR370 pKa = 11.84VQYY373 pKa = 10.32TITRR377 pKa = 11.84VNSQTAYY384 pKa = 10.23QVV386 pKa = 2.96
MM1 pKa = 7.66EE2 pKa = 5.1IARR5 pKa = 11.84TNKK8 pKa = 9.71NSVVKK13 pKa = 8.68YY14 pKa = 9.71VPAAVGAAYY23 pKa = 10.58QMGKK27 pKa = 10.27SIVPYY32 pKa = 10.54APTIVDD38 pKa = 3.15ALGNVVSRR46 pKa = 11.84ATGRR50 pKa = 11.84KK51 pKa = 8.67KK52 pKa = 10.16KK53 pKa = 10.74SKK55 pKa = 10.31GKK57 pKa = 9.45EE58 pKa = 3.73VQNQIVGGIGAIAAPVSITKK78 pKa = 10.12RR79 pKa = 11.84VRR81 pKa = 11.84GMRR84 pKa = 11.84PSFRR88 pKa = 11.84QTKK91 pKa = 8.35GKK93 pKa = 8.67VHH95 pKa = 5.99IVHH98 pKa = 6.85RR99 pKa = 11.84EE100 pKa = 3.74LVTSVINLVGNFRR113 pKa = 11.84VNNNVSAQIGQFRR126 pKa = 11.84INPSNSSLFTWLPTIASNFDD146 pKa = 3.38SYY148 pKa = 11.49RR149 pKa = 11.84FTSIRR154 pKa = 11.84FVYY157 pKa = 10.57VPLCATTEE165 pKa = 4.01TGRR168 pKa = 11.84VSLFWDD174 pKa = 4.23KK175 pKa = 11.29DD176 pKa = 3.79SQDD179 pKa = 3.91PLPVDD184 pKa = 4.13RR185 pKa = 11.84AALSSYY191 pKa = 9.48GHH193 pKa = 6.23SNEE196 pKa = 3.9GPPWAEE202 pKa = 3.7TTLNVPTDD210 pKa = 3.55GKK212 pKa = 10.76QRR214 pKa = 11.84FVTDD218 pKa = 3.13SNTTDD223 pKa = 3.11RR224 pKa = 11.84KK225 pKa = 10.55LVDD228 pKa = 3.99LGQFAFATYY237 pKa = 10.74AGGSNNQIGDD247 pKa = 3.43IYY249 pKa = 11.18VEE251 pKa = 4.09YY252 pKa = 10.57GVEE255 pKa = 4.13FSEE258 pKa = 4.33AQPAGGLTQYY268 pKa = 8.43ITKK271 pKa = 10.37SVGATASTTGPSYY284 pKa = 11.42VVDD287 pKa = 3.87ANINVNATTANVEE300 pKa = 4.27FFSPGTFLITAVVYY314 pKa = 10.53GSTIASPSMAGGNGTLIGDD333 pKa = 4.35LPVVGGSNASIWTCVFSTTGVSTSVPTFTQAGTGLTRR370 pKa = 11.84VQYY373 pKa = 10.32TITRR377 pKa = 11.84VNSQTAYY384 pKa = 10.23QVV386 pKa = 2.96
Molecular weight: 41.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1741 |
152 |
738 |
348.2 |
38.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.203 ± 0.697 | 2.585 ± 0.726 |
4.25 ± 0.409 | 5.686 ± 0.885 |
3.619 ± 0.226 | 7.41 ± 0.768 |
1.666 ± 0.278 | 5.342 ± 0.522 |
5.342 ± 0.682 | 9.133 ± 1.569 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.757 ± 0.68 | 3.331 ± 0.862 |
4.71 ± 0.175 | 3.389 ± 0.717 |
6.433 ± 0.669 | 8.099 ± 0.826 |
5.744 ± 1.442 | 9.075 ± 0.606 |
1.321 ± 0.164 | 3.848 ± 0.463 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
