
Wheat leaf yellowing-associated virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Polerovirus; unclassified Polerovirus
Average proteome isoelectric point is 7.53
Get precalculated fractions of proteins
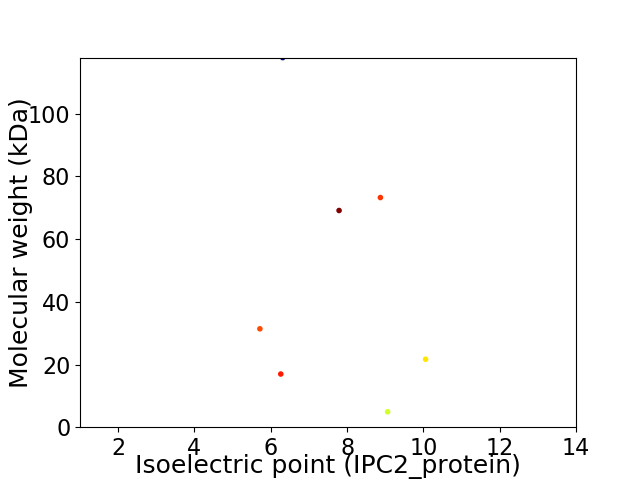
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A220T7N1|A0A220T7N1_9LUTE Readthrough protein OS=Wheat leaf yellowing-associated virus OX=2019445 GN=WLYaV_gp5 PE=3 SV=1
MM1 pKa = 7.48LNAEE5 pKa = 3.9ITAEE9 pKa = 4.1GFVFTGPATSAPATHH24 pKa = 6.94EE25 pKa = 4.15DD26 pKa = 3.75VEE28 pKa = 4.78HH29 pKa = 6.66FLVTYY34 pKa = 10.23RR35 pKa = 11.84RR36 pKa = 11.84SLFLWIHH43 pKa = 5.6EE44 pKa = 4.46CSPFDD49 pKa = 4.08TIRR52 pKa = 11.84VIHH55 pKa = 6.73KK56 pKa = 9.85YY57 pKa = 10.99GSFHH61 pKa = 6.41KK62 pKa = 10.13WGLAVRR68 pKa = 11.84AYY70 pKa = 10.55LCAFIPIILLPGTTINAEE88 pKa = 4.41LISADD93 pKa = 3.72GIFRR97 pKa = 11.84KK98 pKa = 9.95SIYY101 pKa = 9.87VWLARR106 pKa = 11.84FKK108 pKa = 9.66QHH110 pKa = 5.69YY111 pKa = 9.24RR112 pKa = 11.84IASRR116 pKa = 11.84RR117 pKa = 11.84GRR119 pKa = 11.84CLIYY123 pKa = 10.27HH124 pKa = 6.44KK125 pKa = 10.84QPDD128 pKa = 3.73GSFEE132 pKa = 4.83SNLWRR137 pKa = 11.84AVWAPEE143 pKa = 3.89LQSVEE148 pKa = 3.76RR149 pKa = 11.84FKK151 pKa = 11.11RR152 pKa = 11.84ITEE155 pKa = 4.08RR156 pKa = 11.84GLEE159 pKa = 4.12SFKK162 pKa = 10.68TALNLDD168 pKa = 3.77ILPLEE173 pKa = 4.31RR174 pKa = 11.84VHH176 pKa = 7.99KK177 pKa = 9.93IICGKK182 pKa = 9.32PALDD186 pKa = 4.67DD187 pKa = 4.26DD188 pKa = 4.37QVISILSWYY197 pKa = 10.26DD198 pKa = 4.06DD199 pKa = 3.56ILDD202 pKa = 4.88GYY204 pKa = 10.76DD205 pKa = 3.63SEE207 pKa = 5.32DD208 pKa = 3.62NPVEE212 pKa = 4.04TRR214 pKa = 11.84AGMRR218 pKa = 11.84LFGCGFYY225 pKa = 11.01NDD227 pKa = 4.61LLDD230 pKa = 3.87VEE232 pKa = 4.95KK233 pKa = 11.04DD234 pKa = 3.64LLIHH238 pKa = 6.36SFMVGWSSTHH248 pKa = 6.28SPFYY252 pKa = 11.21DD253 pKa = 3.25NTLVGQTADD262 pKa = 3.56QIRR265 pKa = 11.84IWGEE269 pKa = 3.29QWRR272 pKa = 4.06
MM1 pKa = 7.48LNAEE5 pKa = 3.9ITAEE9 pKa = 4.1GFVFTGPATSAPATHH24 pKa = 6.94EE25 pKa = 4.15DD26 pKa = 3.75VEE28 pKa = 4.78HH29 pKa = 6.66FLVTYY34 pKa = 10.23RR35 pKa = 11.84RR36 pKa = 11.84SLFLWIHH43 pKa = 5.6EE44 pKa = 4.46CSPFDD49 pKa = 4.08TIRR52 pKa = 11.84VIHH55 pKa = 6.73KK56 pKa = 9.85YY57 pKa = 10.99GSFHH61 pKa = 6.41KK62 pKa = 10.13WGLAVRR68 pKa = 11.84AYY70 pKa = 10.55LCAFIPIILLPGTTINAEE88 pKa = 4.41LISADD93 pKa = 3.72GIFRR97 pKa = 11.84KK98 pKa = 9.95SIYY101 pKa = 9.87VWLARR106 pKa = 11.84FKK108 pKa = 9.66QHH110 pKa = 5.69YY111 pKa = 9.24RR112 pKa = 11.84IASRR116 pKa = 11.84RR117 pKa = 11.84GRR119 pKa = 11.84CLIYY123 pKa = 10.27HH124 pKa = 6.44KK125 pKa = 10.84QPDD128 pKa = 3.73GSFEE132 pKa = 4.83SNLWRR137 pKa = 11.84AVWAPEE143 pKa = 3.89LQSVEE148 pKa = 3.76RR149 pKa = 11.84FKK151 pKa = 11.11RR152 pKa = 11.84ITEE155 pKa = 4.08RR156 pKa = 11.84GLEE159 pKa = 4.12SFKK162 pKa = 10.68TALNLDD168 pKa = 3.77ILPLEE173 pKa = 4.31RR174 pKa = 11.84VHH176 pKa = 7.99KK177 pKa = 9.93IICGKK182 pKa = 9.32PALDD186 pKa = 4.67DD187 pKa = 4.26DD188 pKa = 4.37QVISILSWYY197 pKa = 10.26DD198 pKa = 4.06DD199 pKa = 3.56ILDD202 pKa = 4.88GYY204 pKa = 10.76DD205 pKa = 3.63SEE207 pKa = 5.32DD208 pKa = 3.62NPVEE212 pKa = 4.04TRR214 pKa = 11.84AGMRR218 pKa = 11.84LFGCGFYY225 pKa = 11.01NDD227 pKa = 4.61LLDD230 pKa = 3.87VEE232 pKa = 4.95KK233 pKa = 11.04DD234 pKa = 3.64LLIHH238 pKa = 6.36SFMVGWSSTHH248 pKa = 6.28SPFYY252 pKa = 11.21DD253 pKa = 3.25NTLVGQTADD262 pKa = 3.56QIRR265 pKa = 11.84IWGEE269 pKa = 3.29QWRR272 pKa = 4.06
Molecular weight: 31.39 kDa
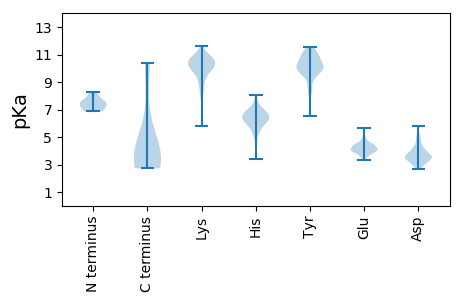
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A220T7N5|A0A220T7N5_9LUTE Movement protein OS=Wheat leaf yellowing-associated virus OX=2019445 GN=WLYaV_gp7 PE=3 SV=1
MM1 pKa = 6.87NTGGNRR7 pKa = 11.84TRR9 pKa = 11.84RR10 pKa = 11.84NARR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84ANRR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84STRR23 pKa = 11.84PVVVVRR29 pKa = 11.84PTPKK33 pKa = 8.88PRR35 pKa = 11.84RR36 pKa = 11.84VRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84ASAGGGAVRR50 pKa = 11.84GPGGRR55 pKa = 11.84SNRR58 pKa = 11.84EE59 pKa = 3.59VFTFTVDD66 pKa = 3.41DD67 pKa = 4.61LKK69 pKa = 11.6ANSSGILKK77 pKa = 9.99FGPNLSQYY85 pKa = 11.26AAFNNGILKK94 pKa = 10.24AYY96 pKa = 9.77HH97 pKa = 6.5EE98 pKa = 4.42YY99 pKa = 10.71KK100 pKa = 9.38ITSLTIQYY108 pKa = 9.32NSCSSSTTSGAIALEE123 pKa = 4.21VDD125 pKa = 4.38TSCSQTTTGSKK136 pKa = 9.73IVSFPVKK143 pKa = 10.49SNTRR147 pKa = 11.84KK148 pKa = 7.85TFPTSFIRR156 pKa = 11.84GKK158 pKa = 10.76DD159 pKa = 3.74FVTTTADD166 pKa = 4.14QFWLLYY172 pKa = 9.95KK173 pKa = 11.05GNGDD177 pKa = 3.49SSLAGQFVCRR187 pKa = 11.84FEE189 pKa = 5.38CQFQNPKK196 pKa = 10.4
MM1 pKa = 6.87NTGGNRR7 pKa = 11.84TRR9 pKa = 11.84RR10 pKa = 11.84NARR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84ANRR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84STRR23 pKa = 11.84PVVVVRR29 pKa = 11.84PTPKK33 pKa = 8.88PRR35 pKa = 11.84RR36 pKa = 11.84VRR38 pKa = 11.84RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84ASAGGGAVRR50 pKa = 11.84GPGGRR55 pKa = 11.84SNRR58 pKa = 11.84EE59 pKa = 3.59VFTFTVDD66 pKa = 3.41DD67 pKa = 4.61LKK69 pKa = 11.6ANSSGILKK77 pKa = 9.99FGPNLSQYY85 pKa = 11.26AAFNNGILKK94 pKa = 10.24AYY96 pKa = 9.77HH97 pKa = 6.5EE98 pKa = 4.42YY99 pKa = 10.71KK100 pKa = 9.38ITSLTIQYY108 pKa = 9.32NSCSSSTTSGAIALEE123 pKa = 4.21VDD125 pKa = 4.38TSCSQTTTGSKK136 pKa = 9.73IVSFPVKK143 pKa = 10.49SNTRR147 pKa = 11.84KK148 pKa = 7.85TFPTSFIRR156 pKa = 11.84GKK158 pKa = 10.76DD159 pKa = 3.74FVTTTADD166 pKa = 4.14QFWLLYY172 pKa = 9.95KK173 pKa = 11.05GNGDD177 pKa = 3.49SSLAGQFVCRR187 pKa = 11.84FEE189 pKa = 5.38CQFQNPKK196 pKa = 10.4
Molecular weight: 21.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3004 |
44 |
1045 |
429.1 |
47.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.691 ± 0.275 | 1.731 ± 0.234 |
4.128 ± 0.403 | 6.092 ± 0.65 |
3.395 ± 0.441 | 7.057 ± 0.489 |
2.031 ± 0.402 | 4.727 ± 0.589 |
6.059 ± 0.591 | 8.722 ± 0.902 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.197 ± 0.447 | 4.294 ± 0.591 |
5.726 ± 0.472 | 3.862 ± 0.568 |
5.659 ± 1.081 | 9.92 ± 0.709 |
6.192 ± 0.636 | 5.659 ± 0.226 |
2.031 ± 0.376 | 3.795 ± 0.193 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
