
Sutterella sp. CAG:521
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Sutterellaceae; Sutterella; environmental samples
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
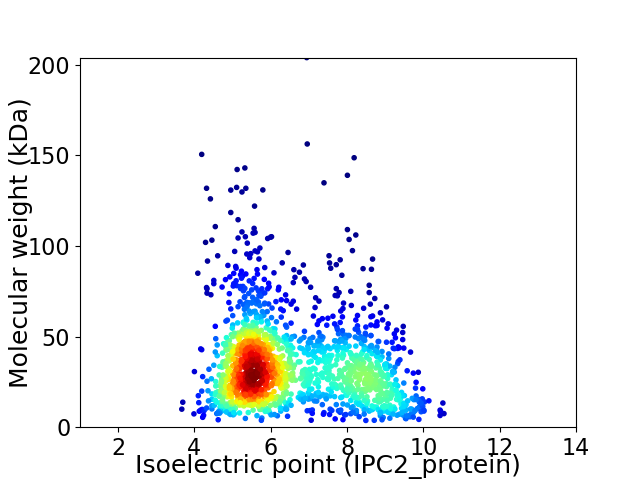
Virtual 2D-PAGE plot for 1632 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R7KGP2|R7KGP2_9BURK Outer membrane autotransporter barrel domain protein OS=Sutterella sp. CAG:521 OX=1262977 GN=BN692_01007 PE=4 SV=1
MM1 pKa = 7.62SSFKK5 pKa = 10.62KK6 pKa = 10.55SSFVMALMVAVGSAQAASVEE26 pKa = 4.55VYY28 pKa = 9.43GTVDD32 pKa = 2.82TGLLYY37 pKa = 10.1THH39 pKa = 7.23EE40 pKa = 4.75SSDD43 pKa = 3.67SKK45 pKa = 11.66VNNEE49 pKa = 4.47KK50 pKa = 10.39KK51 pKa = 10.11HH52 pKa = 5.72EE53 pKa = 4.28SADD56 pKa = 3.41NWGVASGTNTASVIGIRR73 pKa = 11.84GMEE76 pKa = 4.64NISDD80 pKa = 4.03DD81 pKa = 3.96LSVGFVLEE89 pKa = 4.12NSFNSDD95 pKa = 3.09DD96 pKa = 3.96GSFAEE101 pKa = 4.94EE102 pKa = 3.45NTLFDD107 pKa = 4.87KK108 pKa = 10.66EE109 pKa = 4.13AQLFVTSSFGTVSLGRR125 pKa = 11.84MGALTAGEE133 pKa = 4.2GTYY136 pKa = 10.4DD137 pKa = 2.83IFMVNGDD144 pKa = 4.17AMDD147 pKa = 4.35GGYY150 pKa = 10.81ADD152 pKa = 5.0YY153 pKa = 10.86IGAGYY158 pKa = 10.13WMDD161 pKa = 3.4RR162 pKa = 11.84GIYY165 pKa = 10.37DD166 pKa = 4.04NMISFQSPEE175 pKa = 3.92FAGITAFAQYY185 pKa = 11.25SFGTSGDD192 pKa = 3.59DD193 pKa = 3.33MVPSRR198 pKa = 11.84DD199 pKa = 3.4KK200 pKa = 10.94EE201 pKa = 3.93RR202 pKa = 11.84YY203 pKa = 8.04AALGATFSAGNFSAVAVVDD222 pKa = 3.62TVMKK226 pKa = 10.55NRR228 pKa = 11.84QTVTGYY234 pKa = 11.16DD235 pKa = 3.42KK236 pKa = 11.36AIDD239 pKa = 3.62DD240 pKa = 4.52AMAFSIGANYY250 pKa = 10.95DD251 pKa = 3.29MGFMKK256 pKa = 10.32PFIGFQYY263 pKa = 9.8GQHH266 pKa = 5.27EE267 pKa = 4.28QMIGGVPVVEE277 pKa = 4.17TDD279 pKa = 3.11EE280 pKa = 4.45TEE282 pKa = 4.51SYY284 pKa = 11.26VGDD287 pKa = 3.73LDD289 pKa = 4.28GYY291 pKa = 10.46AVAVGSAFPIWGGEE305 pKa = 4.08LQVSVYY311 pKa = 10.62YY312 pKa = 11.05ADD314 pKa = 4.58GDD316 pKa = 4.06GKK318 pKa = 10.94AYY320 pKa = 10.17GYY322 pKa = 10.72DD323 pKa = 3.43QDD325 pKa = 4.99ATTQSPKK332 pKa = 9.15VSKK335 pKa = 10.53MNVKK339 pKa = 10.25RR340 pKa = 11.84YY341 pKa = 9.45GVALFHH347 pKa = 6.92NYY349 pKa = 9.05EE350 pKa = 3.95LSKK353 pKa = 10.37RR354 pKa = 11.84TSLYY358 pKa = 10.83AGIGYY363 pKa = 9.38DD364 pKa = 3.41YY365 pKa = 11.3QEE367 pKa = 4.24YY368 pKa = 10.8DD369 pKa = 3.09EE370 pKa = 6.01SYY372 pKa = 10.49VNSSEE377 pKa = 4.61KK378 pKa = 10.33EE379 pKa = 3.82HH380 pKa = 7.19SEE382 pKa = 3.8QDD384 pKa = 3.59SVQVGVGLVHH394 pKa = 6.97NFF396 pKa = 3.08
MM1 pKa = 7.62SSFKK5 pKa = 10.62KK6 pKa = 10.55SSFVMALMVAVGSAQAASVEE26 pKa = 4.55VYY28 pKa = 9.43GTVDD32 pKa = 2.82TGLLYY37 pKa = 10.1THH39 pKa = 7.23EE40 pKa = 4.75SSDD43 pKa = 3.67SKK45 pKa = 11.66VNNEE49 pKa = 4.47KK50 pKa = 10.39KK51 pKa = 10.11HH52 pKa = 5.72EE53 pKa = 4.28SADD56 pKa = 3.41NWGVASGTNTASVIGIRR73 pKa = 11.84GMEE76 pKa = 4.64NISDD80 pKa = 4.03DD81 pKa = 3.96LSVGFVLEE89 pKa = 4.12NSFNSDD95 pKa = 3.09DD96 pKa = 3.96GSFAEE101 pKa = 4.94EE102 pKa = 3.45NTLFDD107 pKa = 4.87KK108 pKa = 10.66EE109 pKa = 4.13AQLFVTSSFGTVSLGRR125 pKa = 11.84MGALTAGEE133 pKa = 4.2GTYY136 pKa = 10.4DD137 pKa = 2.83IFMVNGDD144 pKa = 4.17AMDD147 pKa = 4.35GGYY150 pKa = 10.81ADD152 pKa = 5.0YY153 pKa = 10.86IGAGYY158 pKa = 10.13WMDD161 pKa = 3.4RR162 pKa = 11.84GIYY165 pKa = 10.37DD166 pKa = 4.04NMISFQSPEE175 pKa = 3.92FAGITAFAQYY185 pKa = 11.25SFGTSGDD192 pKa = 3.59DD193 pKa = 3.33MVPSRR198 pKa = 11.84DD199 pKa = 3.4KK200 pKa = 10.94EE201 pKa = 3.93RR202 pKa = 11.84YY203 pKa = 8.04AALGATFSAGNFSAVAVVDD222 pKa = 3.62TVMKK226 pKa = 10.55NRR228 pKa = 11.84QTVTGYY234 pKa = 11.16DD235 pKa = 3.42KK236 pKa = 11.36AIDD239 pKa = 3.62DD240 pKa = 4.52AMAFSIGANYY250 pKa = 10.95DD251 pKa = 3.29MGFMKK256 pKa = 10.32PFIGFQYY263 pKa = 9.8GQHH266 pKa = 5.27EE267 pKa = 4.28QMIGGVPVVEE277 pKa = 4.17TDD279 pKa = 3.11EE280 pKa = 4.45TEE282 pKa = 4.51SYY284 pKa = 11.26VGDD287 pKa = 3.73LDD289 pKa = 4.28GYY291 pKa = 10.46AVAVGSAFPIWGGEE305 pKa = 4.08LQVSVYY311 pKa = 10.62YY312 pKa = 11.05ADD314 pKa = 4.58GDD316 pKa = 4.06GKK318 pKa = 10.94AYY320 pKa = 10.17GYY322 pKa = 10.72DD323 pKa = 3.43QDD325 pKa = 4.99ATTQSPKK332 pKa = 9.15VSKK335 pKa = 10.53MNVKK339 pKa = 10.25RR340 pKa = 11.84YY341 pKa = 9.45GVALFHH347 pKa = 6.92NYY349 pKa = 9.05EE350 pKa = 3.95LSKK353 pKa = 10.37RR354 pKa = 11.84TSLYY358 pKa = 10.83AGIGYY363 pKa = 9.38DD364 pKa = 3.41YY365 pKa = 11.3QEE367 pKa = 4.24YY368 pKa = 10.8DD369 pKa = 3.09EE370 pKa = 6.01SYY372 pKa = 10.49VNSSEE377 pKa = 4.61KK378 pKa = 10.33EE379 pKa = 3.82HH380 pKa = 7.19SEE382 pKa = 3.8QDD384 pKa = 3.59SVQVGVGLVHH394 pKa = 6.97NFF396 pKa = 3.08
Molecular weight: 42.78 kDa
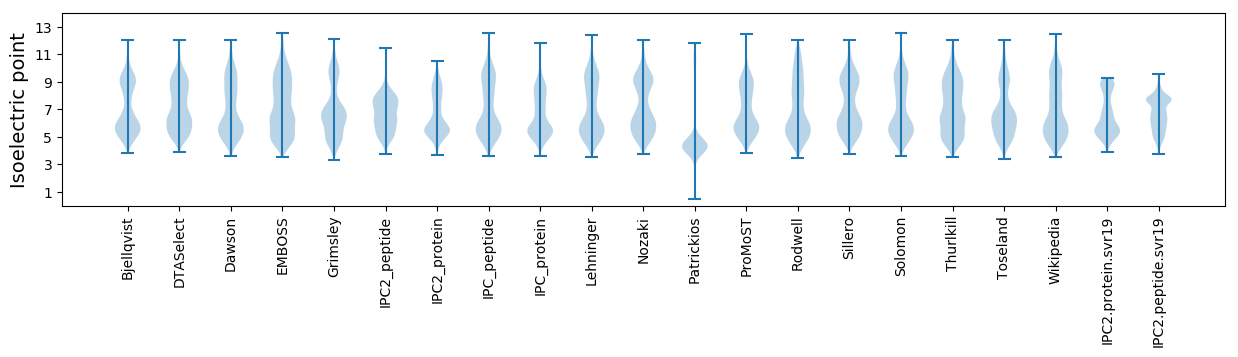
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R7KM13|R7KM13_9BURK Alginate O-acetylation protein OS=Sutterella sp. CAG:521 OX=1262977 GN=BN692_01596 PE=3 SV=1
MM1 pKa = 7.58ALVKK5 pKa = 10.36LKK7 pKa = 8.84PTSAGRR13 pKa = 11.84RR14 pKa = 11.84HH15 pKa = 5.4AVKK18 pKa = 10.3VVSPEE23 pKa = 3.82LHH25 pKa = 6.13KK26 pKa = 11.05GKK28 pKa = 9.55PFAALLEE35 pKa = 4.4KK36 pKa = 10.46KK37 pKa = 10.45NRR39 pKa = 11.84IAGRR43 pKa = 11.84NNNGHH48 pKa = 4.98ITCRR52 pKa = 11.84HH53 pKa = 5.84KK54 pKa = 11.32GGGHH58 pKa = 5.61KK59 pKa = 9.77KK60 pKa = 9.89AYY62 pKa = 10.11RR63 pKa = 11.84IIDD66 pKa = 3.77FKK68 pKa = 11.1RR69 pKa = 11.84NKK71 pKa = 10.05DD72 pKa = 3.47GVPARR77 pKa = 11.84VEE79 pKa = 3.82RR80 pKa = 11.84LEE82 pKa = 3.89YY83 pKa = 10.73DD84 pKa = 3.78PNRR87 pKa = 11.84SAHH90 pKa = 6.08IALVLYY96 pKa = 10.73ADD98 pKa = 3.86GEE100 pKa = 4.19RR101 pKa = 11.84RR102 pKa = 11.84YY103 pKa = 10.3IIAPKK108 pKa = 9.91GLTVGAEE115 pKa = 3.93LMNGQEE121 pKa = 4.29APIKK125 pKa = 10.44VGNALPLRR133 pKa = 11.84NIPIGSTIHH142 pKa = 6.51CIEE145 pKa = 4.15MKK147 pKa = 9.67PGKK150 pKa = 9.77GAQLVRR156 pKa = 11.84SAGSFAQLIARR167 pKa = 11.84EE168 pKa = 4.17GEE170 pKa = 4.19YY171 pKa = 10.86AQVRR175 pKa = 11.84LRR177 pKa = 11.84SGEE180 pKa = 3.57VRR182 pKa = 11.84YY183 pKa = 10.05IHH185 pKa = 5.66VEE187 pKa = 3.84CRR189 pKa = 11.84ATIGMVGNEE198 pKa = 3.98EE199 pKa = 3.78NSLRR203 pKa = 11.84EE204 pKa = 4.01LGKK207 pKa = 10.63AGAKK211 pKa = 8.19RR212 pKa = 11.84WRR214 pKa = 11.84GIRR217 pKa = 11.84PTVRR221 pKa = 11.84GVAMNPVDD229 pKa = 3.85HH230 pKa = 6.82PHH232 pKa = 6.7GGGEE236 pKa = 4.05GRR238 pKa = 11.84TGTGRR243 pKa = 11.84APVTPWGQLTKK254 pKa = 10.73GYY256 pKa = 7.72RR257 pKa = 11.84TRR259 pKa = 11.84NNKK262 pKa = 8.36RR263 pKa = 11.84TDD265 pKa = 2.86AMIVQRR271 pKa = 11.84RR272 pKa = 11.84NRR274 pKa = 11.84KK275 pKa = 8.66
MM1 pKa = 7.58ALVKK5 pKa = 10.36LKK7 pKa = 8.84PTSAGRR13 pKa = 11.84RR14 pKa = 11.84HH15 pKa = 5.4AVKK18 pKa = 10.3VVSPEE23 pKa = 3.82LHH25 pKa = 6.13KK26 pKa = 11.05GKK28 pKa = 9.55PFAALLEE35 pKa = 4.4KK36 pKa = 10.46KK37 pKa = 10.45NRR39 pKa = 11.84IAGRR43 pKa = 11.84NNNGHH48 pKa = 4.98ITCRR52 pKa = 11.84HH53 pKa = 5.84KK54 pKa = 11.32GGGHH58 pKa = 5.61KK59 pKa = 9.77KK60 pKa = 9.89AYY62 pKa = 10.11RR63 pKa = 11.84IIDD66 pKa = 3.77FKK68 pKa = 11.1RR69 pKa = 11.84NKK71 pKa = 10.05DD72 pKa = 3.47GVPARR77 pKa = 11.84VEE79 pKa = 3.82RR80 pKa = 11.84LEE82 pKa = 3.89YY83 pKa = 10.73DD84 pKa = 3.78PNRR87 pKa = 11.84SAHH90 pKa = 6.08IALVLYY96 pKa = 10.73ADD98 pKa = 3.86GEE100 pKa = 4.19RR101 pKa = 11.84RR102 pKa = 11.84YY103 pKa = 10.3IIAPKK108 pKa = 9.91GLTVGAEE115 pKa = 3.93LMNGQEE121 pKa = 4.29APIKK125 pKa = 10.44VGNALPLRR133 pKa = 11.84NIPIGSTIHH142 pKa = 6.51CIEE145 pKa = 4.15MKK147 pKa = 9.67PGKK150 pKa = 9.77GAQLVRR156 pKa = 11.84SAGSFAQLIARR167 pKa = 11.84EE168 pKa = 4.17GEE170 pKa = 4.19YY171 pKa = 10.86AQVRR175 pKa = 11.84LRR177 pKa = 11.84SGEE180 pKa = 3.57VRR182 pKa = 11.84YY183 pKa = 10.05IHH185 pKa = 5.66VEE187 pKa = 3.84CRR189 pKa = 11.84ATIGMVGNEE198 pKa = 3.98EE199 pKa = 3.78NSLRR203 pKa = 11.84EE204 pKa = 4.01LGKK207 pKa = 10.63AGAKK211 pKa = 8.19RR212 pKa = 11.84WRR214 pKa = 11.84GIRR217 pKa = 11.84PTVRR221 pKa = 11.84GVAMNPVDD229 pKa = 3.85HH230 pKa = 6.82PHH232 pKa = 6.7GGGEE236 pKa = 4.05GRR238 pKa = 11.84TGTGRR243 pKa = 11.84APVTPWGQLTKK254 pKa = 10.73GYY256 pKa = 7.72RR257 pKa = 11.84TRR259 pKa = 11.84NNKK262 pKa = 8.36RR263 pKa = 11.84TDD265 pKa = 2.86AMIVQRR271 pKa = 11.84RR272 pKa = 11.84NRR274 pKa = 11.84KK275 pKa = 8.66
Molecular weight: 30.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
526924 |
32 |
1847 |
322.9 |
35.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.282 ± 0.061 | 1.261 ± 0.025 |
5.345 ± 0.049 | 6.322 ± 0.067 |
4.298 ± 0.045 | 7.113 ± 0.066 |
2.048 ± 0.028 | 6.27 ± 0.046 |
5.61 ± 0.055 | 10.038 ± 0.08 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.779 ± 0.034 | 3.707 ± 0.044 |
4.048 ± 0.035 | 3.698 ± 0.039 |
5.334 ± 0.055 | 5.959 ± 0.049 |
5.449 ± 0.04 | 7.423 ± 0.05 |
1.222 ± 0.022 | 2.793 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
