
Infectious laryngotracheitis virus (ILTV) (Gallid herpesvirus 1)
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Alphaherpesvirinae; Iltovirus
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
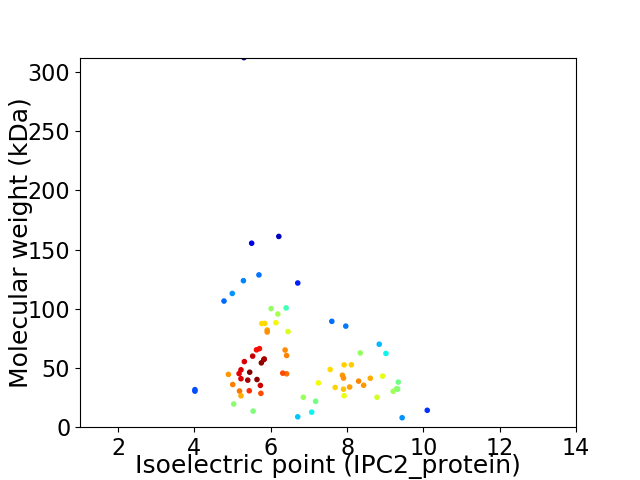
Virtual 2D-PAGE plot for 75 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
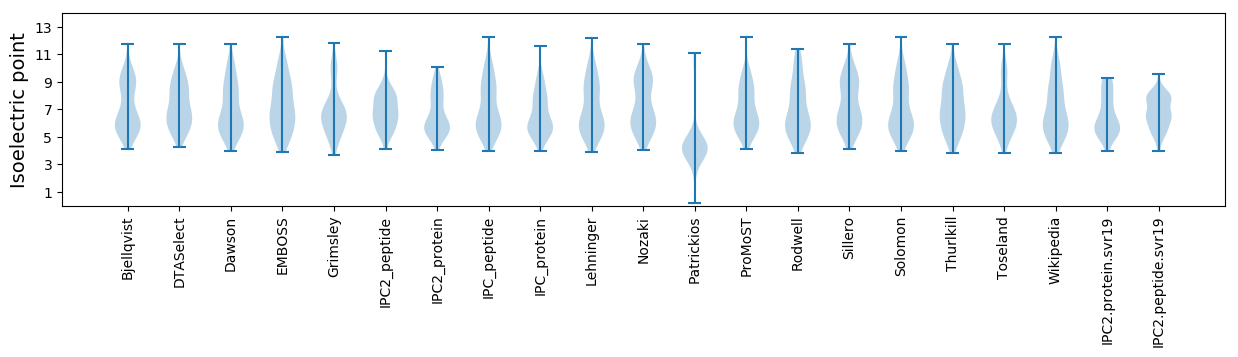
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q67644|Q67644_ILTV Envelope glycoprotein D OS=Infectious laryngotracheitis virus OX=10386 GN=US6 PE=4 SV=1
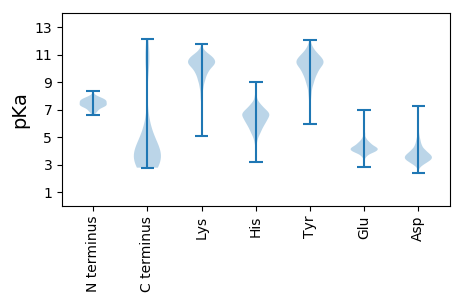
MM1 pKa = 7.57SGFSNIGSIATVSLVCSLLCASVLGAPVLDD31 pKa = 4.33GLEE34 pKa = 4.13SSPFPFGGKK43 pKa = 9.7IIAQACNRR51 pKa = 11.84TTIEE55 pKa = 4.04VTVPWSDD62 pKa = 3.39YY63 pKa = 10.44SGRR66 pKa = 11.84TEE68 pKa = 4.1GVSVEE73 pKa = 4.37VKK75 pKa = 9.29WFYY78 pKa = 11.61GNSNPEE84 pKa = 3.97SFVFGVDD91 pKa = 3.2SEE93 pKa = 4.72TGSGHH98 pKa = 6.8EE99 pKa = 4.98DD100 pKa = 4.98LSTCWALIHH109 pKa = 6.21NLNASVCRR117 pKa = 11.84ASDD120 pKa = 3.18AGIPDD125 pKa = 4.04FDD127 pKa = 4.05KK128 pKa = 11.16QCEE131 pKa = 4.12KK132 pKa = 10.0VQRR135 pKa = 11.84RR136 pKa = 11.84LRR138 pKa = 11.84SGVEE142 pKa = 3.48LGSYY146 pKa = 10.34VSGNGSLVLYY156 pKa = 9.96PGMYY160 pKa = 10.18DD161 pKa = 3.14AGIYY165 pKa = 9.99AYY167 pKa = 10.0QLSVGGKK174 pKa = 8.44GYY176 pKa = 9.07TGSVYY181 pKa = 10.77LDD183 pKa = 3.26VGPNPGCHH191 pKa = 6.13DD192 pKa = 3.6QYY194 pKa = 11.66GYY196 pKa = 9.51TYY198 pKa = 11.07YY199 pKa = 11.28SLADD203 pKa = 3.66EE204 pKa = 5.21ASDD207 pKa = 3.74LSSYY211 pKa = 11.19DD212 pKa = 3.37VASPEE217 pKa = 3.9LDD219 pKa = 3.2GPMEE223 pKa = 4.39EE224 pKa = 5.73DD225 pKa = 3.7YY226 pKa = 11.61SNCLDD231 pKa = 4.08MPPLRR236 pKa = 11.84PWTTVCSHH244 pKa = 6.89DD245 pKa = 4.71VEE247 pKa = 4.43EE248 pKa = 4.8QEE250 pKa = 4.34NATDD254 pKa = 5.25EE255 pKa = 4.67LYY257 pKa = 10.97LWDD260 pKa = 4.54EE261 pKa = 4.48EE262 pKa = 4.81CAGPLDD268 pKa = 4.09EE269 pKa = 5.5YY270 pKa = 11.28VDD272 pKa = 3.96EE273 pKa = 5.45RR274 pKa = 11.84SEE276 pKa = 4.07TMPRR280 pKa = 11.84MVVFSPPSTLQQQ292 pKa = 3.16
MM1 pKa = 7.57SGFSNIGSIATVSLVCSLLCASVLGAPVLDD31 pKa = 4.33GLEE34 pKa = 4.13SSPFPFGGKK43 pKa = 9.7IIAQACNRR51 pKa = 11.84TTIEE55 pKa = 4.04VTVPWSDD62 pKa = 3.39YY63 pKa = 10.44SGRR66 pKa = 11.84TEE68 pKa = 4.1GVSVEE73 pKa = 4.37VKK75 pKa = 9.29WFYY78 pKa = 11.61GNSNPEE84 pKa = 3.97SFVFGVDD91 pKa = 3.2SEE93 pKa = 4.72TGSGHH98 pKa = 6.8EE99 pKa = 4.98DD100 pKa = 4.98LSTCWALIHH109 pKa = 6.21NLNASVCRR117 pKa = 11.84ASDD120 pKa = 3.18AGIPDD125 pKa = 4.04FDD127 pKa = 4.05KK128 pKa = 11.16QCEE131 pKa = 4.12KK132 pKa = 10.0VQRR135 pKa = 11.84RR136 pKa = 11.84LRR138 pKa = 11.84SGVEE142 pKa = 3.48LGSYY146 pKa = 10.34VSGNGSLVLYY156 pKa = 9.96PGMYY160 pKa = 10.18DD161 pKa = 3.14AGIYY165 pKa = 9.99AYY167 pKa = 10.0QLSVGGKK174 pKa = 8.44GYY176 pKa = 9.07TGSVYY181 pKa = 10.77LDD183 pKa = 3.26VGPNPGCHH191 pKa = 6.13DD192 pKa = 3.6QYY194 pKa = 11.66GYY196 pKa = 9.51TYY198 pKa = 11.07YY199 pKa = 11.28SLADD203 pKa = 3.66EE204 pKa = 5.21ASDD207 pKa = 3.74LSSYY211 pKa = 11.19DD212 pKa = 3.37VASPEE217 pKa = 3.9LDD219 pKa = 3.2GPMEE223 pKa = 4.39EE224 pKa = 5.73DD225 pKa = 3.7YY226 pKa = 11.61SNCLDD231 pKa = 4.08MPPLRR236 pKa = 11.84PWTTVCSHH244 pKa = 6.89DD245 pKa = 4.71VEE247 pKa = 4.43EE248 pKa = 4.8QEE250 pKa = 4.34NATDD254 pKa = 5.25EE255 pKa = 4.67LYY257 pKa = 10.97LWDD260 pKa = 4.54EE261 pKa = 4.48EE262 pKa = 4.81CAGPLDD268 pKa = 4.09EE269 pKa = 5.5YY270 pKa = 11.28VDD272 pKa = 3.96EE273 pKa = 5.45RR274 pKa = 11.84SEE276 pKa = 4.07TMPRR280 pKa = 11.84MVVFSPPSTLQQQ292 pKa = 3.16
Molecular weight: 31.7 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q98111|Q98111_ILTV Isoform of F5B4W6 Protein LORF2 OS=Infectious laryngotracheitis virus OX=10386 GN=UL[-1] PE=4 SV=2
MM1 pKa = 7.99KK2 pKa = 10.27YY3 pKa = 10.02GQAAIATDD11 pKa = 3.44MDD13 pKa = 4.61FARR16 pKa = 11.84MSRR19 pKa = 11.84QPPRR23 pKa = 11.84KK24 pKa = 9.47NSRR27 pKa = 11.84CSSARR32 pKa = 11.84TAALQGNGYY41 pKa = 9.91CFLPKK46 pKa = 9.78SEE48 pKa = 5.24KK49 pKa = 10.71LPEE52 pKa = 4.22LPSRR56 pKa = 11.84HH57 pKa = 5.81FEE59 pKa = 4.11TRR61 pKa = 11.84FSSLLPPAAAKK72 pKa = 10.22
MM1 pKa = 7.99KK2 pKa = 10.27YY3 pKa = 10.02GQAAIATDD11 pKa = 3.44MDD13 pKa = 4.61FARR16 pKa = 11.84MSRR19 pKa = 11.84QPPRR23 pKa = 11.84KK24 pKa = 9.47NSRR27 pKa = 11.84CSSARR32 pKa = 11.84TAALQGNGYY41 pKa = 9.91CFLPKK46 pKa = 9.78SEE48 pKa = 5.24KK49 pKa = 10.71LPEE52 pKa = 4.22LPSRR56 pKa = 11.84HH57 pKa = 5.81FEE59 pKa = 4.11TRR61 pKa = 11.84FSSLLPPAAAKK72 pKa = 10.22
Molecular weight: 7.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
39631 |
72 |
2784 |
528.4 |
59.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.979 ± 0.203 | 2.162 ± 0.121 |
5.19 ± 0.162 | 6.399 ± 0.212 |
4.401 ± 0.172 | 5.617 ± 0.228 |
2.233 ± 0.078 | 5.526 ± 0.205 |
4.464 ± 0.236 | 9.702 ± 0.203 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.162 ± 0.086 | 3.739 ± 0.112 |
5.708 ± 0.286 | 3.101 ± 0.119 |
6.543 ± 0.259 | 8.567 ± 0.192 |
6.192 ± 0.256 | 5.955 ± 0.149 |
1.153 ± 0.066 | 3.207 ± 0.107 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
