
Cellulomonas sp. NEAU-YY56
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Cellulomonadaceae; Cellulomonas; unclassified Cellulomonas
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
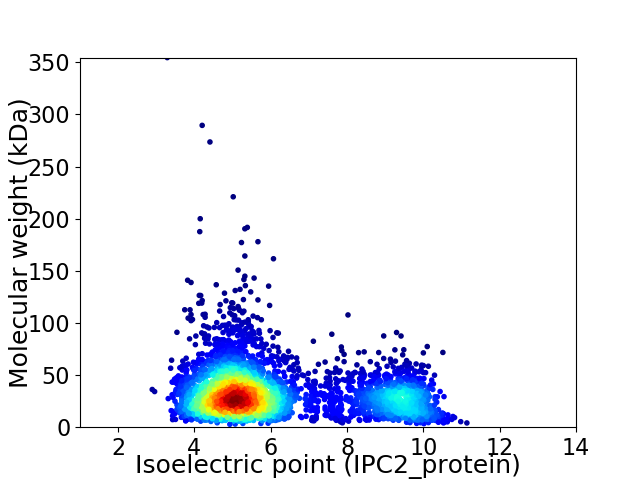
Virtual 2D-PAGE plot for 4017 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3M2J461|A0A3M2J461_9CELL Alpha-D-glucose phosphate-specific phosphoglucomutase OS=Cellulomonas sp. NEAU-YY56 OX=2483352 GN=EBM89_12640 PE=3 SV=1
MM1 pKa = 7.01TAVPHH6 pKa = 5.37GRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84GIVTGAGLVATALVLAACGGSGDD33 pKa = 4.49DD34 pKa = 4.32SSDD37 pKa = 3.22GGGDD41 pKa = 3.51GGSGGALLIGTTDD54 pKa = 4.02KK55 pKa = 10.49ITTIDD60 pKa = 3.49PAGSYY65 pKa = 11.17DD66 pKa = 3.54NGSFAVMNQIYY77 pKa = 8.82PFLMNTPYY85 pKa = 10.85GSPDD89 pKa = 3.56VEE91 pKa = 3.89PDD93 pKa = 3.01IAEE96 pKa = 4.25SAEE99 pKa = 4.09FTSPTEE105 pKa = 3.79YY106 pKa = 10.53TVTLKK111 pKa = 10.98EE112 pKa = 4.14GLTFANGNDD121 pKa = 3.63LTSSDD126 pKa = 3.75VKK128 pKa = 10.88FSFDD132 pKa = 2.97RR133 pKa = 11.84QLAIFEE139 pKa = 4.56SGADD143 pKa = 3.78DD144 pKa = 4.27GNGPGSLLYY153 pKa = 11.01NLDD156 pKa = 3.53SVAAPDD162 pKa = 4.09DD163 pKa = 3.73LTVVFTLKK171 pKa = 10.97SPDD174 pKa = 3.65DD175 pKa = 3.57QVFPQILSSPVGPIVDD191 pKa = 3.83EE192 pKa = 4.73DD193 pKa = 4.16VFAADD198 pKa = 4.7ALTPDD203 pKa = 3.73AEE205 pKa = 4.49IVDD208 pKa = 4.35GQAFAGPYY216 pKa = 10.22VLTDD220 pKa = 3.23YY221 pKa = 10.96AQNDD225 pKa = 4.17LLSYY229 pKa = 8.41TANPDD234 pKa = 3.47YY235 pKa = 10.82QGLLGAPKK243 pKa = 9.69TEE245 pKa = 4.66EE246 pKa = 4.11INVQYY251 pKa = 9.51YY252 pKa = 10.09TDD254 pKa = 3.67ASNLKK259 pKa = 10.73LEE261 pKa = 4.43VQQGTVDD268 pKa = 3.28VAFRR272 pKa = 11.84SLSATDD278 pKa = 3.76IEE280 pKa = 5.27DD281 pKa = 4.16LRR283 pKa = 11.84GDD285 pKa = 4.08DD286 pKa = 3.5NVKK289 pKa = 10.33VVDD292 pKa = 4.44GPGGEE297 pKa = 3.65IRR299 pKa = 11.84YY300 pKa = 8.28ITFNFNTQPYY310 pKa = 9.66GATTPEE316 pKa = 4.22ADD318 pKa = 3.52PAKK321 pKa = 10.57ALAVRR326 pKa = 11.84QAVADD331 pKa = 4.63LIDD334 pKa = 3.98RR335 pKa = 11.84EE336 pKa = 4.31EE337 pKa = 4.55IADD340 pKa = 3.64QVYY343 pKa = 10.41KK344 pKa = 10.16GTYY347 pKa = 7.53TPLYY351 pKa = 9.34SYY353 pKa = 10.97VPEE356 pKa = 4.62GLTGANEE363 pKa = 4.08ALKK366 pKa = 10.81GLYY369 pKa = 10.38GDD371 pKa = 4.56GEE373 pKa = 4.58GGADD377 pKa = 3.34ADD379 pKa = 4.19KK380 pKa = 11.05AAEE383 pKa = 4.1TLEE386 pKa = 4.26AAGVEE391 pKa = 4.6TPVQLSLQYY400 pKa = 11.43SNDD403 pKa = 3.17HH404 pKa = 6.36YY405 pKa = 11.18GPSSGEE411 pKa = 3.74EE412 pKa = 3.59YY413 pKa = 11.33ALIKK417 pKa = 10.67DD418 pKa = 3.81QLEE421 pKa = 4.46STGLFAVDD429 pKa = 4.19LQTTEE434 pKa = 3.56WVQYY438 pKa = 11.09AKK440 pKa = 10.96DD441 pKa = 3.58RR442 pKa = 11.84TADD445 pKa = 3.41VYY447 pKa = 10.63PAYY450 pKa = 10.26QLGWFPDD457 pKa = 3.67YY458 pKa = 11.48SDD460 pKa = 4.11ADD462 pKa = 3.57NYY464 pKa = 8.96LTPFFLTDD472 pKa = 3.18NFLGNHH478 pKa = 6.09YY479 pKa = 11.16DD480 pKa = 3.59NAEE483 pKa = 4.06VNDD486 pKa = 5.44LILQQAVTTDD496 pKa = 3.3TAEE499 pKa = 3.89RR500 pKa = 11.84TALIGEE506 pKa = 4.25IQDD509 pKa = 4.82AVAQDD514 pKa = 4.57LSTVPYY520 pKa = 10.3LQGAQVAVVGADD532 pKa = 3.2VTGAEE537 pKa = 4.4DD538 pKa = 3.51TLDD541 pKa = 3.3ASFKK545 pKa = 10.38FRR547 pKa = 11.84YY548 pKa = 8.66GALAIGG554 pKa = 3.91
MM1 pKa = 7.01TAVPHH6 pKa = 5.37GRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84GIVTGAGLVATALVLAACGGSGDD33 pKa = 4.49DD34 pKa = 4.32SSDD37 pKa = 3.22GGGDD41 pKa = 3.51GGSGGALLIGTTDD54 pKa = 4.02KK55 pKa = 10.49ITTIDD60 pKa = 3.49PAGSYY65 pKa = 11.17DD66 pKa = 3.54NGSFAVMNQIYY77 pKa = 8.82PFLMNTPYY85 pKa = 10.85GSPDD89 pKa = 3.56VEE91 pKa = 3.89PDD93 pKa = 3.01IAEE96 pKa = 4.25SAEE99 pKa = 4.09FTSPTEE105 pKa = 3.79YY106 pKa = 10.53TVTLKK111 pKa = 10.98EE112 pKa = 4.14GLTFANGNDD121 pKa = 3.63LTSSDD126 pKa = 3.75VKK128 pKa = 10.88FSFDD132 pKa = 2.97RR133 pKa = 11.84QLAIFEE139 pKa = 4.56SGADD143 pKa = 3.78DD144 pKa = 4.27GNGPGSLLYY153 pKa = 11.01NLDD156 pKa = 3.53SVAAPDD162 pKa = 4.09DD163 pKa = 3.73LTVVFTLKK171 pKa = 10.97SPDD174 pKa = 3.65DD175 pKa = 3.57QVFPQILSSPVGPIVDD191 pKa = 3.83EE192 pKa = 4.73DD193 pKa = 4.16VFAADD198 pKa = 4.7ALTPDD203 pKa = 3.73AEE205 pKa = 4.49IVDD208 pKa = 4.35GQAFAGPYY216 pKa = 10.22VLTDD220 pKa = 3.23YY221 pKa = 10.96AQNDD225 pKa = 4.17LLSYY229 pKa = 8.41TANPDD234 pKa = 3.47YY235 pKa = 10.82QGLLGAPKK243 pKa = 9.69TEE245 pKa = 4.66EE246 pKa = 4.11INVQYY251 pKa = 9.51YY252 pKa = 10.09TDD254 pKa = 3.67ASNLKK259 pKa = 10.73LEE261 pKa = 4.43VQQGTVDD268 pKa = 3.28VAFRR272 pKa = 11.84SLSATDD278 pKa = 3.76IEE280 pKa = 5.27DD281 pKa = 4.16LRR283 pKa = 11.84GDD285 pKa = 4.08DD286 pKa = 3.5NVKK289 pKa = 10.33VVDD292 pKa = 4.44GPGGEE297 pKa = 3.65IRR299 pKa = 11.84YY300 pKa = 8.28ITFNFNTQPYY310 pKa = 9.66GATTPEE316 pKa = 4.22ADD318 pKa = 3.52PAKK321 pKa = 10.57ALAVRR326 pKa = 11.84QAVADD331 pKa = 4.63LIDD334 pKa = 3.98RR335 pKa = 11.84EE336 pKa = 4.31EE337 pKa = 4.55IADD340 pKa = 3.64QVYY343 pKa = 10.41KK344 pKa = 10.16GTYY347 pKa = 7.53TPLYY351 pKa = 9.34SYY353 pKa = 10.97VPEE356 pKa = 4.62GLTGANEE363 pKa = 4.08ALKK366 pKa = 10.81GLYY369 pKa = 10.38GDD371 pKa = 4.56GEE373 pKa = 4.58GGADD377 pKa = 3.34ADD379 pKa = 4.19KK380 pKa = 11.05AAEE383 pKa = 4.1TLEE386 pKa = 4.26AAGVEE391 pKa = 4.6TPVQLSLQYY400 pKa = 11.43SNDD403 pKa = 3.17HH404 pKa = 6.36YY405 pKa = 11.18GPSSGEE411 pKa = 3.74EE412 pKa = 3.59YY413 pKa = 11.33ALIKK417 pKa = 10.67DD418 pKa = 3.81QLEE421 pKa = 4.46STGLFAVDD429 pKa = 4.19LQTTEE434 pKa = 3.56WVQYY438 pKa = 11.09AKK440 pKa = 10.96DD441 pKa = 3.58RR442 pKa = 11.84TADD445 pKa = 3.41VYY447 pKa = 10.63PAYY450 pKa = 10.26QLGWFPDD457 pKa = 3.67YY458 pKa = 11.48SDD460 pKa = 4.11ADD462 pKa = 3.57NYY464 pKa = 8.96LTPFFLTDD472 pKa = 3.18NFLGNHH478 pKa = 6.09YY479 pKa = 11.16DD480 pKa = 3.59NAEE483 pKa = 4.06VNDD486 pKa = 5.44LILQQAVTTDD496 pKa = 3.3TAEE499 pKa = 3.89RR500 pKa = 11.84TALIGEE506 pKa = 4.25IQDD509 pKa = 4.82AVAQDD514 pKa = 4.57LSTVPYY520 pKa = 10.3LQGAQVAVVGADD532 pKa = 3.2VTGAEE537 pKa = 4.4DD538 pKa = 3.51TLDD541 pKa = 3.3ASFKK545 pKa = 10.38FRR547 pKa = 11.84YY548 pKa = 8.66GALAIGG554 pKa = 3.91
Molecular weight: 58.72 kDa
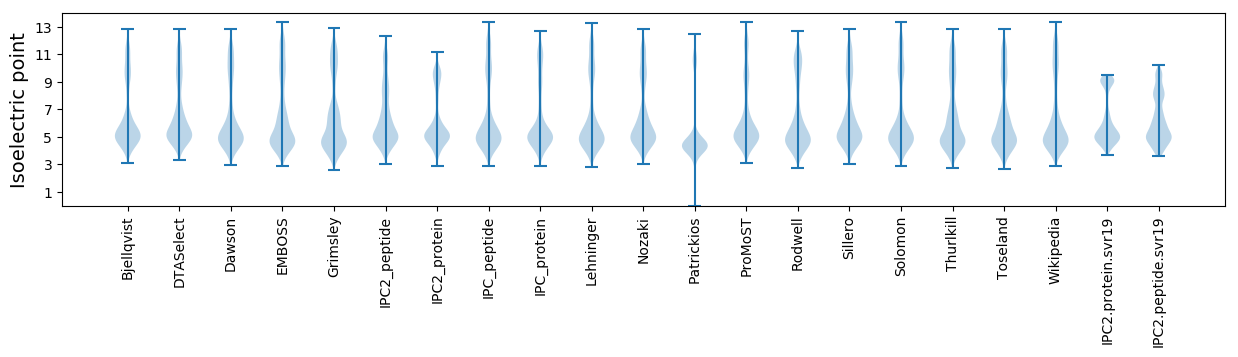
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3M2JFS1|A0A3M2JFS1_9CELL DUF885 domain-containing protein OS=Cellulomonas sp. NEAU-YY56 OX=2483352 GN=EBM89_07975 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1294718 |
29 |
3745 |
322.3 |
34.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.894 ± 0.064 | 0.527 ± 0.01 |
6.587 ± 0.04 | 5.04 ± 0.035 |
2.422 ± 0.023 | 9.61 ± 0.04 |
2.035 ± 0.023 | 2.683 ± 0.028 |
1.204 ± 0.02 | 10.459 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.509 ± 0.016 | 1.369 ± 0.018 |
6.048 ± 0.037 | 2.626 ± 0.022 |
7.91 ± 0.052 | 4.739 ± 0.03 |
6.54 ± 0.046 | 10.486 ± 0.048 |
1.529 ± 0.016 | 1.783 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
