
Candidatus Nitrosocosmicus arcticus
Taxonomy: cellular organisms; Archaea; TACK group; Thaumarchaeota; Nitrososphaeria; Nitrososphaerales; Nitrososphaeraceae; Candidatus Nitrosocosmicus
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
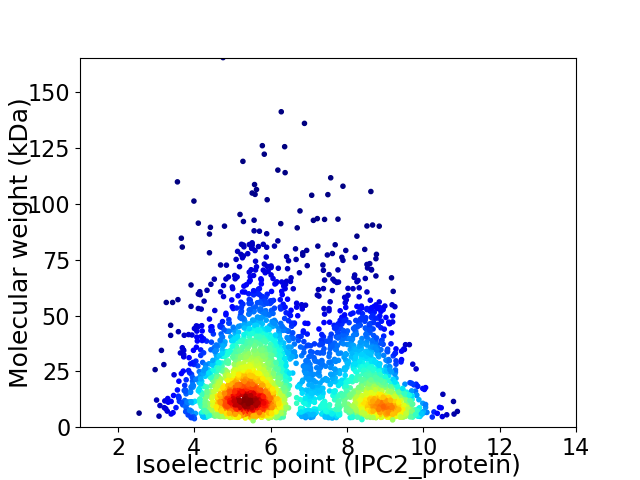
Virtual 2D-PAGE plot for 3069 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A557SVZ9|A0A557SVZ9_9ARCH Putative ester cyclase OS=Candidatus Nitrosocosmicus arcticus OX=2035267 GN=NARC_60163 PE=4 SV=1
MM1 pKa = 9.01VMIKK5 pKa = 10.39YY6 pKa = 9.81VLIPIFVVITLFTTGLVEE24 pKa = 4.07NSFSVEE30 pKa = 3.72LRR32 pKa = 11.84DD33 pKa = 4.09LKK35 pKa = 10.41TIISNDD41 pKa = 3.38EE42 pKa = 3.74ATKK45 pKa = 10.24EE46 pKa = 3.9YY47 pKa = 11.07AVIPNQEE54 pKa = 4.47LLVIPLTDD62 pKa = 3.39INSIAYY68 pKa = 8.32YY69 pKa = 10.05QLNTFDD75 pKa = 4.39QSYY78 pKa = 8.13TFQNVTVLEE87 pKa = 4.34GVVYY91 pKa = 10.35EE92 pKa = 4.74DD93 pKa = 4.72KK94 pKa = 11.06KK95 pKa = 11.28SNNDD99 pKa = 3.09NGATSGDD106 pKa = 4.08LDD108 pKa = 4.52CSDD111 pKa = 3.15VAARR115 pKa = 11.84NFLVGSSDD123 pKa = 3.31SYY125 pKa = 11.95NFDD128 pKa = 3.59RR129 pKa = 11.84DD130 pKa = 3.29GDD132 pKa = 4.39GIGCEE137 pKa = 3.89SSNYY141 pKa = 8.78YY142 pKa = 10.59NDD144 pKa = 3.66YY145 pKa = 10.52NSLYY149 pKa = 11.03AVTSDD154 pKa = 3.49NNSRR158 pKa = 11.84CPDD161 pKa = 3.31GFHH164 pKa = 7.33RR165 pKa = 11.84SPSGDD170 pKa = 3.56CEE172 pKa = 4.0RR173 pKa = 11.84VTDD176 pKa = 3.68TSGMEE181 pKa = 3.74RR182 pKa = 11.84CPDD185 pKa = 3.33GFHH188 pKa = 7.27RR189 pKa = 11.84SPSGDD194 pKa = 3.52CEE196 pKa = 3.84RR197 pKa = 11.84VTDD200 pKa = 5.11NNDD203 pKa = 3.05NDD205 pKa = 4.45KK206 pKa = 11.29GSNPCTPIEE215 pKa = 3.96ASKK218 pKa = 11.32DD219 pKa = 3.47HH220 pKa = 6.65TCTSGSEE227 pKa = 4.26GIDD230 pKa = 3.15YY231 pKa = 10.28SKK233 pKa = 11.17PNNNDD238 pKa = 2.79NDD240 pKa = 3.91ASNNDD245 pKa = 3.58YY246 pKa = 11.7DD247 pKa = 6.23NIDD250 pKa = 3.39NWTDD254 pKa = 3.5PPAEE258 pKa = 4.19EE259 pKa = 4.19TTEE262 pKa = 4.23GNGVEE267 pKa = 4.29NEE269 pKa = 3.96EE270 pKa = 5.35SGQQEE275 pKa = 4.14NNEE278 pKa = 4.07NTEE281 pKa = 4.0EE282 pKa = 4.23QPNNDD287 pKa = 4.08EE288 pKa = 4.9GSSSSDD294 pKa = 3.27EE295 pKa = 4.41GSSSSDD301 pKa = 3.27EE302 pKa = 4.41GSSSSDD308 pKa = 3.13EE309 pKa = 4.12GSSDD313 pKa = 3.61NNN315 pKa = 3.5
MM1 pKa = 9.01VMIKK5 pKa = 10.39YY6 pKa = 9.81VLIPIFVVITLFTTGLVEE24 pKa = 4.07NSFSVEE30 pKa = 3.72LRR32 pKa = 11.84DD33 pKa = 4.09LKK35 pKa = 10.41TIISNDD41 pKa = 3.38EE42 pKa = 3.74ATKK45 pKa = 10.24EE46 pKa = 3.9YY47 pKa = 11.07AVIPNQEE54 pKa = 4.47LLVIPLTDD62 pKa = 3.39INSIAYY68 pKa = 8.32YY69 pKa = 10.05QLNTFDD75 pKa = 4.39QSYY78 pKa = 8.13TFQNVTVLEE87 pKa = 4.34GVVYY91 pKa = 10.35EE92 pKa = 4.74DD93 pKa = 4.72KK94 pKa = 11.06KK95 pKa = 11.28SNNDD99 pKa = 3.09NGATSGDD106 pKa = 4.08LDD108 pKa = 4.52CSDD111 pKa = 3.15VAARR115 pKa = 11.84NFLVGSSDD123 pKa = 3.31SYY125 pKa = 11.95NFDD128 pKa = 3.59RR129 pKa = 11.84DD130 pKa = 3.29GDD132 pKa = 4.39GIGCEE137 pKa = 3.89SSNYY141 pKa = 8.78YY142 pKa = 10.59NDD144 pKa = 3.66YY145 pKa = 10.52NSLYY149 pKa = 11.03AVTSDD154 pKa = 3.49NNSRR158 pKa = 11.84CPDD161 pKa = 3.31GFHH164 pKa = 7.33RR165 pKa = 11.84SPSGDD170 pKa = 3.56CEE172 pKa = 4.0RR173 pKa = 11.84VTDD176 pKa = 3.68TSGMEE181 pKa = 3.74RR182 pKa = 11.84CPDD185 pKa = 3.33GFHH188 pKa = 7.27RR189 pKa = 11.84SPSGDD194 pKa = 3.52CEE196 pKa = 3.84RR197 pKa = 11.84VTDD200 pKa = 5.11NNDD203 pKa = 3.05NDD205 pKa = 4.45KK206 pKa = 11.29GSNPCTPIEE215 pKa = 3.96ASKK218 pKa = 11.32DD219 pKa = 3.47HH220 pKa = 6.65TCTSGSEE227 pKa = 4.26GIDD230 pKa = 3.15YY231 pKa = 10.28SKK233 pKa = 11.17PNNNDD238 pKa = 2.79NDD240 pKa = 3.91ASNNDD245 pKa = 3.58YY246 pKa = 11.7DD247 pKa = 6.23NIDD250 pKa = 3.39NWTDD254 pKa = 3.5PPAEE258 pKa = 4.19EE259 pKa = 4.19TTEE262 pKa = 4.23GNGVEE267 pKa = 4.29NEE269 pKa = 3.96EE270 pKa = 5.35SGQQEE275 pKa = 4.14NNEE278 pKa = 4.07NTEE281 pKa = 4.0EE282 pKa = 4.23QPNNDD287 pKa = 4.08EE288 pKa = 4.9GSSSSDD294 pKa = 3.27EE295 pKa = 4.41GSSSSDD301 pKa = 3.27EE302 pKa = 4.41GSSSSDD308 pKa = 3.13EE309 pKa = 4.12GSSDD313 pKa = 3.61NNN315 pKa = 3.5
Molecular weight: 34.37 kDa
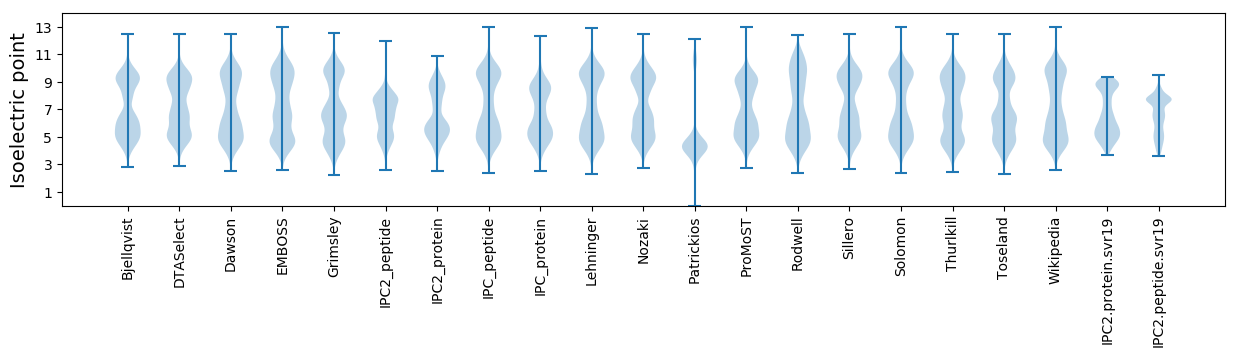
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A557STA8|A0A557STA8_9ARCH Methylenetetrahydrofolate reductase (NAD(P)H) OS=Candidatus Nitrosocosmicus arcticus OX=2035267 GN=metF PE=4 SV=1
MM1 pKa = 7.25KK2 pKa = 9.86RR3 pKa = 11.84APPPPPLTPLTAGMKK18 pKa = 9.71RR19 pKa = 11.84APPPPPLTPLTAGMKK34 pKa = 9.95KK35 pKa = 10.2ALPLMSLLRR44 pKa = 11.84HH45 pKa = 6.14PNLLIQLAQIRR56 pKa = 11.84RR57 pKa = 11.84KK58 pKa = 8.79LTTWRR63 pKa = 11.84KK64 pKa = 8.7KK65 pKa = 9.72IRR67 pKa = 11.84AISINKK73 pKa = 9.4LVDD76 pKa = 3.09NAFFANTGIFGTKK89 pKa = 9.82NNSKK93 pKa = 9.42MPRR96 pKa = 11.84PTNIKK101 pKa = 10.13VNSS104 pKa = 3.69
MM1 pKa = 7.25KK2 pKa = 9.86RR3 pKa = 11.84APPPPPLTPLTAGMKK18 pKa = 9.71RR19 pKa = 11.84APPPPPLTPLTAGMKK34 pKa = 9.95KK35 pKa = 10.2ALPLMSLLRR44 pKa = 11.84HH45 pKa = 6.14PNLLIQLAQIRR56 pKa = 11.84RR57 pKa = 11.84KK58 pKa = 8.79LTTWRR63 pKa = 11.84KK64 pKa = 8.7KK65 pKa = 9.72IRR67 pKa = 11.84AISINKK73 pKa = 9.4LVDD76 pKa = 3.09NAFFANTGIFGTKK89 pKa = 9.82NNSKK93 pKa = 9.42MPRR96 pKa = 11.84PTNIKK101 pKa = 10.13VNSS104 pKa = 3.69
Molecular weight: 11.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
668652 |
28 |
1529 |
217.9 |
24.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.961 ± 0.053 | 1.049 ± 0.022 |
5.726 ± 0.04 | 6.215 ± 0.045 |
4.571 ± 0.037 | 6.054 ± 0.052 |
1.806 ± 0.021 | 9.62 ± 0.055 |
7.598 ± 0.049 | 9.121 ± 0.055 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.462 ± 0.024 | 6.635 ± 0.055 |
3.624 ± 0.038 | 3.004 ± 0.033 |
3.734 ± 0.037 | 8.03 ± 0.046 |
5.442 ± 0.046 | 5.94 ± 0.039 |
0.844 ± 0.016 | 3.564 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
