
Epirus cherry virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Miaviricetes; Ourlivirales; Botourmiaviridae; Ourmiavirus
Average proteome isoelectric point is 8.24
Get precalculated fractions of proteins
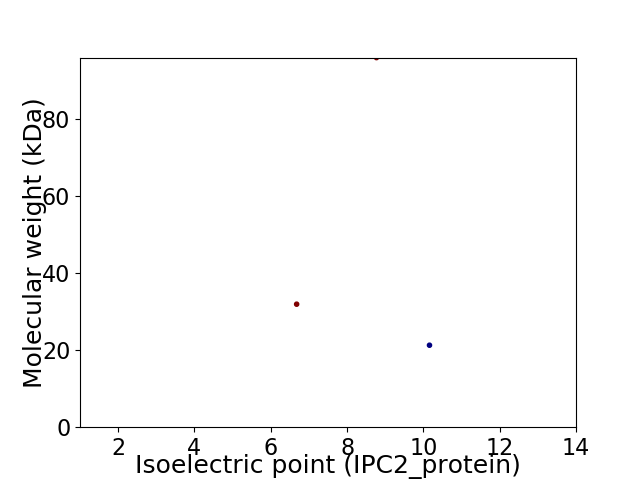
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B3VML0|B3VML0_9VIRU Coat protein OS=Epirus cherry virus OX=544686 PE=2 SV=1
MM1 pKa = 7.74EE2 pKa = 4.61PTGNNPTVNQPMPVVVAPSHH22 pKa = 5.73SGQMRR27 pKa = 11.84VDD29 pKa = 4.25LSSLQIRR36 pKa = 11.84PPPLEE41 pKa = 3.98GKK43 pKa = 10.16GRR45 pKa = 11.84VVFVTEE51 pKa = 4.09HH52 pKa = 5.45SAYY55 pKa = 10.66DD56 pKa = 3.69GVRR59 pKa = 11.84PLPLIPKK66 pKa = 9.7DD67 pKa = 3.28LAGQLRR73 pKa = 11.84LKK75 pKa = 9.24WHH77 pKa = 6.18EE78 pKa = 3.54KK79 pKa = 10.05RR80 pKa = 11.84YY81 pKa = 8.0PHH83 pKa = 6.99TYY85 pKa = 10.35LAFEE89 pKa = 4.43QIEE92 pKa = 4.57SEE94 pKa = 4.52YY95 pKa = 10.63FPHH98 pKa = 7.56IPEE101 pKa = 4.1RR102 pKa = 11.84SAYY105 pKa = 10.25GEE107 pKa = 4.01LALVDD112 pKa = 3.38TRR114 pKa = 11.84YY115 pKa = 8.59ITDD118 pKa = 3.35EE119 pKa = 4.03VQDD122 pKa = 3.54VDD124 pKa = 3.94EE125 pKa = 5.35EE126 pKa = 4.26EE127 pKa = 4.74MIDD130 pKa = 3.7HH131 pKa = 6.97LFSRR135 pKa = 11.84ALWKK139 pKa = 9.48TPEE142 pKa = 4.26LDD144 pKa = 3.1LGKK147 pKa = 10.62GYY149 pKa = 10.76KK150 pKa = 9.39FVSAVPYY157 pKa = 8.92CIPIHH162 pKa = 6.27ARR164 pKa = 11.84GGQGPDD170 pKa = 3.37LEE172 pKa = 4.66RR173 pKa = 11.84DD174 pKa = 3.02IPIRR178 pKa = 11.84LVPRR182 pKa = 11.84ITRR185 pKa = 11.84TDD187 pKa = 2.9LHH189 pKa = 6.2LASRR193 pKa = 11.84SGTIKK198 pKa = 10.56SALKK202 pKa = 10.1VALSSEE208 pKa = 4.65PIQYY212 pKa = 9.9QRR214 pKa = 11.84IEE216 pKa = 4.37SVSAQTLEE224 pKa = 4.05ARR226 pKa = 11.84NSQITARR233 pKa = 11.84GRR235 pKa = 11.84RR236 pKa = 11.84NLDD239 pKa = 2.87RR240 pKa = 11.84RR241 pKa = 11.84PRR243 pKa = 11.84GGPWPAMEE251 pKa = 5.29RR252 pKa = 11.84SHH254 pKa = 8.0SMRR257 pKa = 11.84SDD259 pKa = 3.16TQIPIEE265 pKa = 4.27HH266 pKa = 7.23AAPPGGGEE274 pKa = 3.96PSRR277 pKa = 11.84TIIMPAGAVV286 pKa = 3.24
MM1 pKa = 7.74EE2 pKa = 4.61PTGNNPTVNQPMPVVVAPSHH22 pKa = 5.73SGQMRR27 pKa = 11.84VDD29 pKa = 4.25LSSLQIRR36 pKa = 11.84PPPLEE41 pKa = 3.98GKK43 pKa = 10.16GRR45 pKa = 11.84VVFVTEE51 pKa = 4.09HH52 pKa = 5.45SAYY55 pKa = 10.66DD56 pKa = 3.69GVRR59 pKa = 11.84PLPLIPKK66 pKa = 9.7DD67 pKa = 3.28LAGQLRR73 pKa = 11.84LKK75 pKa = 9.24WHH77 pKa = 6.18EE78 pKa = 3.54KK79 pKa = 10.05RR80 pKa = 11.84YY81 pKa = 8.0PHH83 pKa = 6.99TYY85 pKa = 10.35LAFEE89 pKa = 4.43QIEE92 pKa = 4.57SEE94 pKa = 4.52YY95 pKa = 10.63FPHH98 pKa = 7.56IPEE101 pKa = 4.1RR102 pKa = 11.84SAYY105 pKa = 10.25GEE107 pKa = 4.01LALVDD112 pKa = 3.38TRR114 pKa = 11.84YY115 pKa = 8.59ITDD118 pKa = 3.35EE119 pKa = 4.03VQDD122 pKa = 3.54VDD124 pKa = 3.94EE125 pKa = 5.35EE126 pKa = 4.26EE127 pKa = 4.74MIDD130 pKa = 3.7HH131 pKa = 6.97LFSRR135 pKa = 11.84ALWKK139 pKa = 9.48TPEE142 pKa = 4.26LDD144 pKa = 3.1LGKK147 pKa = 10.62GYY149 pKa = 10.76KK150 pKa = 9.39FVSAVPYY157 pKa = 8.92CIPIHH162 pKa = 6.27ARR164 pKa = 11.84GGQGPDD170 pKa = 3.37LEE172 pKa = 4.66RR173 pKa = 11.84DD174 pKa = 3.02IPIRR178 pKa = 11.84LVPRR182 pKa = 11.84ITRR185 pKa = 11.84TDD187 pKa = 2.9LHH189 pKa = 6.2LASRR193 pKa = 11.84SGTIKK198 pKa = 10.56SALKK202 pKa = 10.1VALSSEE208 pKa = 4.65PIQYY212 pKa = 9.9QRR214 pKa = 11.84IEE216 pKa = 4.37SVSAQTLEE224 pKa = 4.05ARR226 pKa = 11.84NSQITARR233 pKa = 11.84GRR235 pKa = 11.84RR236 pKa = 11.84NLDD239 pKa = 2.87RR240 pKa = 11.84RR241 pKa = 11.84PRR243 pKa = 11.84GGPWPAMEE251 pKa = 5.29RR252 pKa = 11.84SHH254 pKa = 8.0SMRR257 pKa = 11.84SDD259 pKa = 3.16TQIPIEE265 pKa = 4.27HH266 pKa = 7.23AAPPGGGEE274 pKa = 3.96PSRR277 pKa = 11.84TIIMPAGAVV286 pKa = 3.24
Molecular weight: 31.98 kDa
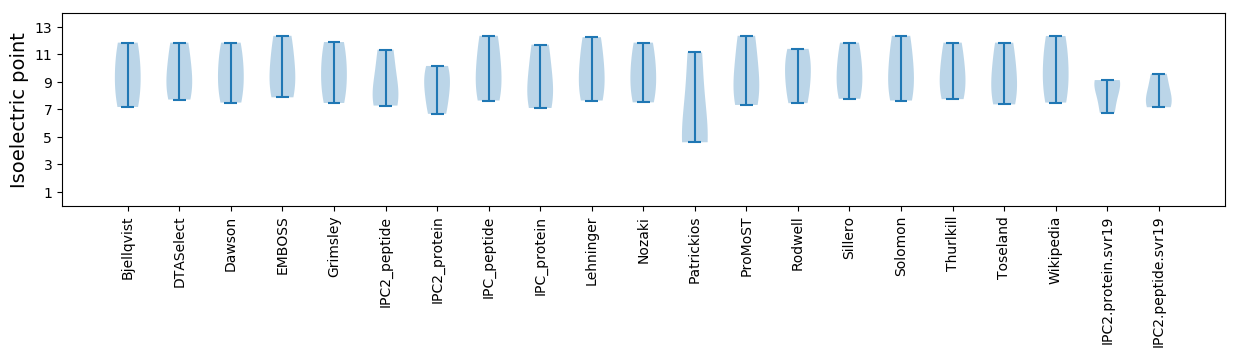
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B3VML0|B3VML0_9VIRU Coat protein OS=Epirus cherry virus OX=544686 PE=2 SV=1
MM1 pKa = 6.92VNRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84GIRR9 pKa = 11.84VRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84NRR15 pKa = 11.84RR16 pKa = 11.84QGTNQQAPIPRR27 pKa = 11.84ISGVRR32 pKa = 11.84SYY34 pKa = 11.24DD35 pKa = 3.27VEE37 pKa = 4.21VFRR40 pKa = 11.84DD41 pKa = 4.22FTFLNRR47 pKa = 11.84RR48 pKa = 11.84NGCAVGQLGPGTGVTQISGLIIPDD72 pKa = 3.72TAISWNIKK80 pKa = 8.58TMSLNPINLVPSTVGGQFLISLVPGPALSGRR111 pKa = 11.84VLADD115 pKa = 3.61LQAIQTGSLMSSPSTLRR132 pKa = 11.84IFPARR137 pKa = 11.84PIPGQPWRR145 pKa = 11.84GSLIQRR151 pKa = 11.84EE152 pKa = 4.45ITRR155 pKa = 11.84SNDD158 pKa = 2.87SRR160 pKa = 11.84VVNPGDD166 pKa = 3.07RR167 pKa = 11.84WGGFAIVSDD176 pKa = 3.76SRR178 pKa = 11.84LLSGLADD185 pKa = 4.42DD186 pKa = 4.77APVWFARR193 pKa = 11.84LSS195 pKa = 3.46
MM1 pKa = 6.92VNRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84GIRR9 pKa = 11.84VRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84NRR15 pKa = 11.84RR16 pKa = 11.84QGTNQQAPIPRR27 pKa = 11.84ISGVRR32 pKa = 11.84SYY34 pKa = 11.24DD35 pKa = 3.27VEE37 pKa = 4.21VFRR40 pKa = 11.84DD41 pKa = 4.22FTFLNRR47 pKa = 11.84RR48 pKa = 11.84NGCAVGQLGPGTGVTQISGLIIPDD72 pKa = 3.72TAISWNIKK80 pKa = 8.58TMSLNPINLVPSTVGGQFLISLVPGPALSGRR111 pKa = 11.84VLADD115 pKa = 3.61LQAIQTGSLMSSPSTLRR132 pKa = 11.84IFPARR137 pKa = 11.84PIPGQPWRR145 pKa = 11.84GSLIQRR151 pKa = 11.84EE152 pKa = 4.45ITRR155 pKa = 11.84SNDD158 pKa = 2.87SRR160 pKa = 11.84VVNPGDD166 pKa = 3.07RR167 pKa = 11.84WGGFAIVSDD176 pKa = 3.76SRR178 pKa = 11.84LLSGLADD185 pKa = 4.42DD186 pKa = 4.77APVWFARR193 pKa = 11.84LSS195 pKa = 3.46
Molecular weight: 21.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1333 |
195 |
852 |
444.3 |
49.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.677 ± 0.286 | 1.2 ± 0.415 |
4.126 ± 0.346 | 5.851 ± 1.333 |
3.001 ± 0.397 | 7.877 ± 0.658 |
2.401 ± 0.671 | 5.326 ± 1.11 |
3.151 ± 0.787 | 10.278 ± 1.034 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.251 ± 0.197 | 2.776 ± 0.655 |
6.977 ± 1.179 | 4.051 ± 0.343 |
9.677 ± 0.733 | 7.727 ± 0.565 |
5.026 ± 0.044 | 7.052 ± 0.257 |
1.575 ± 0.182 | 3.001 ± 0.725 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
