
Wenling crustacean virus 11
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alphacrustrhavirus; Zhejiang alphacrustrhavirus
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
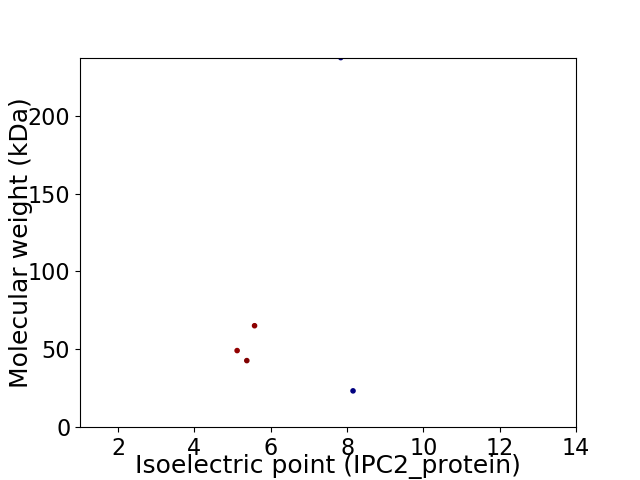
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KN83|A0A1L3KN83_9VIRU Uncharacterized protein OS=Wenling crustacean virus 11 OX=1923480 PE=4 SV=1
MM1 pKa = 7.51ASGVASLEE9 pKa = 3.8RR10 pKa = 11.84LIRR13 pKa = 11.84STDD16 pKa = 2.68ISLNSGRR23 pKa = 11.84GITWNDD29 pKa = 3.01QTLKK33 pKa = 10.8KK34 pKa = 9.81FWKK37 pKa = 9.45RR38 pKa = 11.84HH39 pKa = 4.22YY40 pKa = 10.4AVPNADD46 pKa = 3.86LMKK49 pKa = 10.59SIYY52 pKa = 10.21TDD54 pKa = 3.96FLSAYY59 pKa = 8.94CGKK62 pKa = 10.18FAGFQTGPLIRR73 pKa = 11.84AAILSTLTPPGDD85 pKa = 3.04KK86 pKa = 11.01HH87 pKa = 5.24MFGYY91 pKa = 9.8VFKK94 pKa = 10.54GVVPIYY100 pKa = 10.32DD101 pKa = 3.86PSAPPPPRR109 pKa = 11.84ATPTAKK115 pKa = 9.29TQVPQQSAIQTEE127 pKa = 4.49LSSANTYY134 pKa = 8.97LAKK137 pKa = 10.54DD138 pKa = 3.44NHH140 pKa = 7.02DD141 pKa = 3.51ISSDD145 pKa = 3.86DD146 pKa = 3.74EE147 pKa = 5.87SDD149 pKa = 5.07DD150 pKa = 4.17EE151 pKa = 6.98GDD153 pKa = 4.97DD154 pKa = 4.44EE155 pKa = 6.37PITPPDD161 pKa = 2.97IVGFQEE167 pKa = 4.11TMLKK171 pKa = 10.49YY172 pKa = 10.87IMIAPGNSYY181 pKa = 9.68MVLGFLAMICFRR193 pKa = 11.84IIVRR197 pKa = 11.84DD198 pKa = 3.78PKK200 pKa = 10.84AIRR203 pKa = 11.84LYY205 pKa = 10.21FANQRR210 pKa = 11.84RR211 pKa = 11.84ILALISSCTGVVMRR225 pKa = 11.84TGIPAPTAAFLDD237 pKa = 4.15DD238 pKa = 4.55LVVSIPKK245 pKa = 9.88GQTIASEE252 pKa = 4.33VLADD256 pKa = 3.55MTYY259 pKa = 11.6LLIKK263 pKa = 10.06SGPASPVVRR272 pKa = 11.84FLEE275 pKa = 4.83GGCMVHH281 pKa = 7.6LSGNGLGLIDD291 pKa = 6.29LIDD294 pKa = 3.72RR295 pKa = 11.84VANKK299 pKa = 10.68YY300 pKa = 9.54EE301 pKa = 3.91VTIAMVLSLLLTSRR315 pKa = 11.84SVKK318 pKa = 9.77SVGRR322 pKa = 11.84VVEE325 pKa = 4.31FLVNAPKK332 pKa = 10.29ILSWKK337 pKa = 7.96WSRR340 pKa = 11.84AITDD344 pKa = 3.52RR345 pKa = 11.84AFQDD349 pKa = 4.16LRR351 pKa = 11.84VQNNLTLTGYY361 pKa = 9.91VVALAAEE368 pKa = 4.51ADD370 pKa = 3.79AEE372 pKa = 4.37SEE374 pKa = 3.7IWKK377 pKa = 9.7IRR379 pKa = 11.84ALEE382 pKa = 4.26DD383 pKa = 3.39LPAATKK389 pKa = 10.32AQAIAWAKK397 pKa = 10.15QYY399 pKa = 11.16LILRR403 pKa = 11.84DD404 pKa = 3.89KK405 pKa = 8.84EE406 pKa = 4.6TKK408 pKa = 9.26TSHH411 pKa = 6.54FEE413 pKa = 3.2ASAAYY418 pKa = 9.54RR419 pKa = 11.84RR420 pKa = 11.84VAAGIEE426 pKa = 4.27SEE428 pKa = 4.33EE429 pKa = 4.35EE430 pKa = 3.82EE431 pKa = 4.74SEE433 pKa = 4.81SDD435 pKa = 4.72DD436 pKa = 4.24GDD438 pKa = 3.52ISSDD442 pKa = 3.74EE443 pKa = 5.83DD444 pKa = 3.59DD445 pKa = 5.28APWNN449 pKa = 3.68
MM1 pKa = 7.51ASGVASLEE9 pKa = 3.8RR10 pKa = 11.84LIRR13 pKa = 11.84STDD16 pKa = 2.68ISLNSGRR23 pKa = 11.84GITWNDD29 pKa = 3.01QTLKK33 pKa = 10.8KK34 pKa = 9.81FWKK37 pKa = 9.45RR38 pKa = 11.84HH39 pKa = 4.22YY40 pKa = 10.4AVPNADD46 pKa = 3.86LMKK49 pKa = 10.59SIYY52 pKa = 10.21TDD54 pKa = 3.96FLSAYY59 pKa = 8.94CGKK62 pKa = 10.18FAGFQTGPLIRR73 pKa = 11.84AAILSTLTPPGDD85 pKa = 3.04KK86 pKa = 11.01HH87 pKa = 5.24MFGYY91 pKa = 9.8VFKK94 pKa = 10.54GVVPIYY100 pKa = 10.32DD101 pKa = 3.86PSAPPPPRR109 pKa = 11.84ATPTAKK115 pKa = 9.29TQVPQQSAIQTEE127 pKa = 4.49LSSANTYY134 pKa = 8.97LAKK137 pKa = 10.54DD138 pKa = 3.44NHH140 pKa = 7.02DD141 pKa = 3.51ISSDD145 pKa = 3.86DD146 pKa = 3.74EE147 pKa = 5.87SDD149 pKa = 5.07DD150 pKa = 4.17EE151 pKa = 6.98GDD153 pKa = 4.97DD154 pKa = 4.44EE155 pKa = 6.37PITPPDD161 pKa = 2.97IVGFQEE167 pKa = 4.11TMLKK171 pKa = 10.49YY172 pKa = 10.87IMIAPGNSYY181 pKa = 9.68MVLGFLAMICFRR193 pKa = 11.84IIVRR197 pKa = 11.84DD198 pKa = 3.78PKK200 pKa = 10.84AIRR203 pKa = 11.84LYY205 pKa = 10.21FANQRR210 pKa = 11.84RR211 pKa = 11.84ILALISSCTGVVMRR225 pKa = 11.84TGIPAPTAAFLDD237 pKa = 4.15DD238 pKa = 4.55LVVSIPKK245 pKa = 9.88GQTIASEE252 pKa = 4.33VLADD256 pKa = 3.55MTYY259 pKa = 11.6LLIKK263 pKa = 10.06SGPASPVVRR272 pKa = 11.84FLEE275 pKa = 4.83GGCMVHH281 pKa = 7.6LSGNGLGLIDD291 pKa = 6.29LIDD294 pKa = 3.72RR295 pKa = 11.84VANKK299 pKa = 10.68YY300 pKa = 9.54EE301 pKa = 3.91VTIAMVLSLLLTSRR315 pKa = 11.84SVKK318 pKa = 9.77SVGRR322 pKa = 11.84VVEE325 pKa = 4.31FLVNAPKK332 pKa = 10.29ILSWKK337 pKa = 7.96WSRR340 pKa = 11.84AITDD344 pKa = 3.52RR345 pKa = 11.84AFQDD349 pKa = 4.16LRR351 pKa = 11.84VQNNLTLTGYY361 pKa = 9.91VVALAAEE368 pKa = 4.51ADD370 pKa = 3.79AEE372 pKa = 4.37SEE374 pKa = 3.7IWKK377 pKa = 9.7IRR379 pKa = 11.84ALEE382 pKa = 4.26DD383 pKa = 3.39LPAATKK389 pKa = 10.32AQAIAWAKK397 pKa = 10.15QYY399 pKa = 11.16LILRR403 pKa = 11.84DD404 pKa = 3.89KK405 pKa = 8.84EE406 pKa = 4.6TKK408 pKa = 9.26TSHH411 pKa = 6.54FEE413 pKa = 3.2ASAAYY418 pKa = 9.54RR419 pKa = 11.84RR420 pKa = 11.84VAAGIEE426 pKa = 4.27SEE428 pKa = 4.33EE429 pKa = 4.35EE430 pKa = 3.82EE431 pKa = 4.74SEE433 pKa = 4.81SDD435 pKa = 4.72DD436 pKa = 4.24GDD438 pKa = 3.52ISSDD442 pKa = 3.74EE443 pKa = 5.83DD444 pKa = 3.59DD445 pKa = 5.28APWNN449 pKa = 3.68
Molecular weight: 49.21 kDa
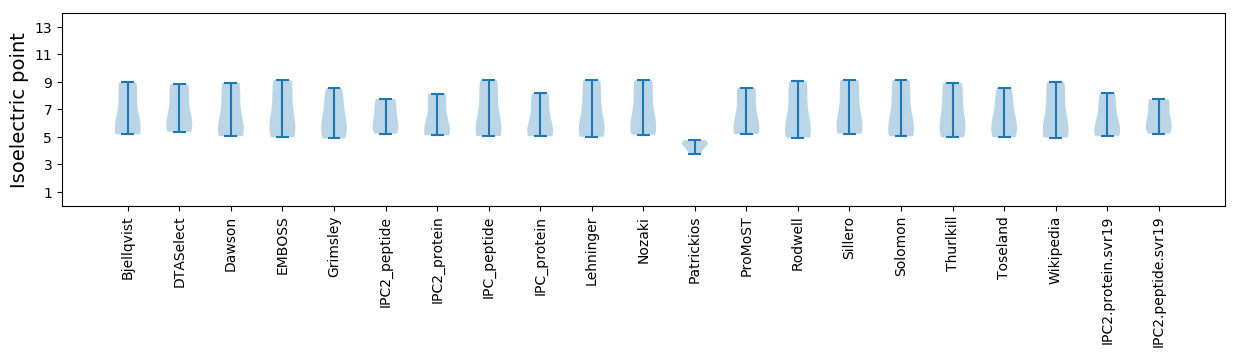
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KN87|A0A1L3KN87_9VIRU Uncharacterized protein OS=Wenling crustacean virus 11 OX=1923480 PE=4 SV=1
MM1 pKa = 8.05LMGRR5 pKa = 11.84VDD7 pKa = 3.63ATVTHH12 pKa = 5.93YY13 pKa = 10.93SEE15 pKa = 4.04YY16 pKa = 7.17TTRR19 pKa = 11.84HH20 pKa = 3.99EE21 pKa = 4.34TIALVILSGLFDD33 pKa = 4.76PYY35 pKa = 11.06LPRR38 pKa = 11.84EE39 pKa = 4.18SLFVLASEE47 pKa = 4.5LSLRR51 pKa = 11.84FSLAEE56 pKa = 4.5SISPDD61 pKa = 3.23CVMGVSDD68 pKa = 4.56GKK70 pKa = 10.08PVVIHH75 pKa = 5.44LQKK78 pKa = 9.92RR79 pKa = 11.84TATVKK84 pKa = 10.45VLYY87 pKa = 10.2ALPDD91 pKa = 3.71EE92 pKa = 5.19GWTHH96 pKa = 5.21TGEE99 pKa = 4.26YY100 pKa = 9.99EE101 pKa = 4.07NTFSAEE107 pKa = 4.23TSCSFGIVSTNCSIHH122 pKa = 5.1ATVRR126 pKa = 11.84QKK128 pKa = 10.82KK129 pKa = 9.76VSDD132 pKa = 3.84VVSKK136 pKa = 11.26KK137 pKa = 10.5MIDD140 pKa = 3.01QGLGYY145 pKa = 10.16RR146 pKa = 11.84RR147 pKa = 11.84LRR149 pKa = 11.84GVGDD153 pKa = 3.35VTRR156 pKa = 11.84GLLINIHH163 pKa = 4.06QTIRR167 pKa = 11.84EE168 pKa = 4.21KK169 pKa = 10.37GTEE172 pKa = 3.69RR173 pKa = 11.84ALKK176 pKa = 9.07HH177 pKa = 5.06VKK179 pKa = 10.13RR180 pKa = 11.84SEE182 pKa = 3.86EE183 pKa = 3.67AASFVGEE190 pKa = 4.16SSLVGPAGGSKK201 pKa = 10.81ALTFLKK207 pKa = 10.9NFVGKK212 pKa = 9.98NN213 pKa = 3.15
MM1 pKa = 8.05LMGRR5 pKa = 11.84VDD7 pKa = 3.63ATVTHH12 pKa = 5.93YY13 pKa = 10.93SEE15 pKa = 4.04YY16 pKa = 7.17TTRR19 pKa = 11.84HH20 pKa = 3.99EE21 pKa = 4.34TIALVILSGLFDD33 pKa = 4.76PYY35 pKa = 11.06LPRR38 pKa = 11.84EE39 pKa = 4.18SLFVLASEE47 pKa = 4.5LSLRR51 pKa = 11.84FSLAEE56 pKa = 4.5SISPDD61 pKa = 3.23CVMGVSDD68 pKa = 4.56GKK70 pKa = 10.08PVVIHH75 pKa = 5.44LQKK78 pKa = 9.92RR79 pKa = 11.84TATVKK84 pKa = 10.45VLYY87 pKa = 10.2ALPDD91 pKa = 3.71EE92 pKa = 5.19GWTHH96 pKa = 5.21TGEE99 pKa = 4.26YY100 pKa = 9.99EE101 pKa = 4.07NTFSAEE107 pKa = 4.23TSCSFGIVSTNCSIHH122 pKa = 5.1ATVRR126 pKa = 11.84QKK128 pKa = 10.82KK129 pKa = 9.76VSDD132 pKa = 3.84VVSKK136 pKa = 11.26KK137 pKa = 10.5MIDD140 pKa = 3.01QGLGYY145 pKa = 10.16RR146 pKa = 11.84RR147 pKa = 11.84LRR149 pKa = 11.84GVGDD153 pKa = 3.35VTRR156 pKa = 11.84GLLINIHH163 pKa = 4.06QTIRR167 pKa = 11.84EE168 pKa = 4.21KK169 pKa = 10.37GTEE172 pKa = 3.69RR173 pKa = 11.84ALKK176 pKa = 9.07HH177 pKa = 5.06VKK179 pKa = 10.13RR180 pKa = 11.84SEE182 pKa = 3.86EE183 pKa = 3.67AASFVGEE190 pKa = 4.16SSLVGPAGGSKK201 pKa = 10.81ALTFLKK207 pKa = 10.9NFVGKK212 pKa = 9.98NN213 pKa = 3.15
Molecular weight: 23.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3742 |
213 |
2106 |
748.4 |
83.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.44 ± 1.216 | 1.336 ± 0.322 |
5.104 ± 0.734 | 6.173 ± 0.535 |
3.554 ± 0.277 | 6.467 ± 0.299 |
2.352 ± 0.286 | 5.746 ± 0.423 |
5.852 ± 0.85 | 10.689 ± 0.679 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.031 ± 0.273 | 2.94 ± 0.177 |
5.158 ± 0.543 | 3.394 ± 0.255 |
5.211 ± 0.125 | 9.621 ± 0.555 |
6.173 ± 0.384 | 6.601 ± 0.475 |
2.138 ± 0.446 | 3.02 ± 0.38 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
