
Anhanga virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Phlebovirus; Anhanga phlebovirus
Average proteome isoelectric point is 6.62
Get precalculated fractions of proteins
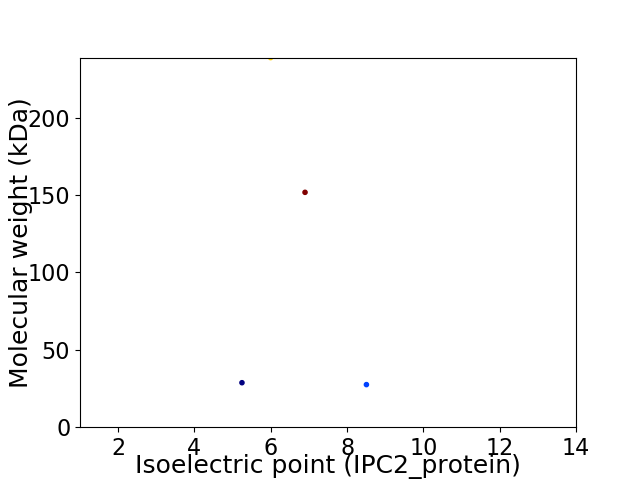
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1S5SHW0|A0A1S5SHW0_9VIRU Nucleoprotein OS=Anhanga virus OX=904722 PE=3 SV=1
MM1 pKa = 7.45LNYY4 pKa = 10.5YY5 pKa = 10.59LFDD8 pKa = 4.27RR9 pKa = 11.84PVVKK13 pKa = 10.38KK14 pKa = 10.59AEE16 pKa = 4.31KK17 pKa = 10.18KK18 pKa = 9.48RR19 pKa = 11.84VVYY22 pKa = 10.08YY23 pKa = 9.07MAHH26 pKa = 5.97NNYY29 pKa = 8.84VSKK32 pKa = 10.59PVACYY37 pKa = 9.89HH38 pKa = 5.55GLEE41 pKa = 4.82FIVDD45 pKa = 3.82HH46 pKa = 6.48FKK48 pKa = 10.98PEE50 pKa = 3.92PSACLDD56 pKa = 3.2FHH58 pKa = 6.73EE59 pKa = 5.7FYY61 pKa = 11.06DD62 pKa = 4.29RR63 pKa = 11.84GYY65 pKa = 11.14LPTLIQGDD73 pKa = 4.32GPSQVRR79 pKa = 11.84SSAPPPDD86 pKa = 4.92GIFEE90 pKa = 4.71FISDD94 pKa = 4.25FSTDD98 pKa = 2.7IFEE101 pKa = 5.43SFDD104 pKa = 3.5EE105 pKa = 4.18MSMLDD110 pKa = 3.48AVRR113 pKa = 11.84WPTGTPSLQFFHH125 pKa = 7.0HH126 pKa = 6.18WRR128 pKa = 11.84NRR130 pKa = 11.84SPGYY134 pKa = 10.27NWTSKK139 pKa = 11.21SMIASEE145 pKa = 4.6LLTATTEE152 pKa = 4.0MCLCCAFPRR161 pKa = 11.84MHH163 pKa = 6.87GFIRR167 pKa = 11.84NIARR171 pKa = 11.84RR172 pKa = 11.84MDD174 pKa = 4.08LDD176 pKa = 4.37LKK178 pKa = 10.54LFPGGDD184 pKa = 3.09IVTEE188 pKa = 3.98ICHH191 pKa = 5.13IQCVKK196 pKa = 9.55MMKK199 pKa = 10.13AALIEE204 pKa = 4.35RR205 pKa = 11.84GEE207 pKa = 4.54DD208 pKa = 3.36KK209 pKa = 10.59PLSPVTEE216 pKa = 5.34LILTAMGEE224 pKa = 4.28YY225 pKa = 9.02TRR227 pKa = 11.84EE228 pKa = 3.74YY229 pKa = 11.15DD230 pKa = 3.8PMLKK234 pKa = 10.49EE235 pKa = 5.2LMDD238 pKa = 3.76EE239 pKa = 4.02AANYY243 pKa = 10.02FDD245 pKa = 4.41EE246 pKa = 4.37TLKK249 pKa = 11.18
MM1 pKa = 7.45LNYY4 pKa = 10.5YY5 pKa = 10.59LFDD8 pKa = 4.27RR9 pKa = 11.84PVVKK13 pKa = 10.38KK14 pKa = 10.59AEE16 pKa = 4.31KK17 pKa = 10.18KK18 pKa = 9.48RR19 pKa = 11.84VVYY22 pKa = 10.08YY23 pKa = 9.07MAHH26 pKa = 5.97NNYY29 pKa = 8.84VSKK32 pKa = 10.59PVACYY37 pKa = 9.89HH38 pKa = 5.55GLEE41 pKa = 4.82FIVDD45 pKa = 3.82HH46 pKa = 6.48FKK48 pKa = 10.98PEE50 pKa = 3.92PSACLDD56 pKa = 3.2FHH58 pKa = 6.73EE59 pKa = 5.7FYY61 pKa = 11.06DD62 pKa = 4.29RR63 pKa = 11.84GYY65 pKa = 11.14LPTLIQGDD73 pKa = 4.32GPSQVRR79 pKa = 11.84SSAPPPDD86 pKa = 4.92GIFEE90 pKa = 4.71FISDD94 pKa = 4.25FSTDD98 pKa = 2.7IFEE101 pKa = 5.43SFDD104 pKa = 3.5EE105 pKa = 4.18MSMLDD110 pKa = 3.48AVRR113 pKa = 11.84WPTGTPSLQFFHH125 pKa = 7.0HH126 pKa = 6.18WRR128 pKa = 11.84NRR130 pKa = 11.84SPGYY134 pKa = 10.27NWTSKK139 pKa = 11.21SMIASEE145 pKa = 4.6LLTATTEE152 pKa = 4.0MCLCCAFPRR161 pKa = 11.84MHH163 pKa = 6.87GFIRR167 pKa = 11.84NIARR171 pKa = 11.84RR172 pKa = 11.84MDD174 pKa = 4.08LDD176 pKa = 4.37LKK178 pKa = 10.54LFPGGDD184 pKa = 3.09IVTEE188 pKa = 3.98ICHH191 pKa = 5.13IQCVKK196 pKa = 9.55MMKK199 pKa = 10.13AALIEE204 pKa = 4.35RR205 pKa = 11.84GEE207 pKa = 4.54DD208 pKa = 3.36KK209 pKa = 10.59PLSPVTEE216 pKa = 5.34LILTAMGEE224 pKa = 4.28YY225 pKa = 9.02TRR227 pKa = 11.84EE228 pKa = 3.74YY229 pKa = 11.15DD230 pKa = 3.8PMLKK234 pKa = 10.49EE235 pKa = 5.2LMDD238 pKa = 3.76EE239 pKa = 4.02AANYY243 pKa = 10.02FDD245 pKa = 4.41EE246 pKa = 4.37TLKK249 pKa = 11.18
Molecular weight: 28.76 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1S5SHW0|A0A1S5SHW0_9VIRU Nucleoprotein OS=Anhanga virus OX=904722 PE=3 SV=1
MM1 pKa = 7.36SFSKK5 pKa = 10.44IALEE9 pKa = 4.16FGGEE13 pKa = 4.23SVDD16 pKa = 3.57VDD18 pKa = 4.95EE19 pKa = 4.6ISKK22 pKa = 9.62WVSEE26 pKa = 4.19FAYY29 pKa = 10.11QGYY32 pKa = 9.44DD33 pKa = 2.98ANRR36 pKa = 11.84VIEE39 pKa = 5.45LIQQYY44 pKa = 8.75GRR46 pKa = 11.84KK47 pKa = 9.01RR48 pKa = 11.84DD49 pKa = 2.99WKK51 pKa = 9.14MDD53 pKa = 3.64VKK55 pKa = 11.06RR56 pKa = 11.84MIVLALTRR64 pKa = 11.84GNKK67 pKa = 7.63PDD69 pKa = 3.45KK70 pKa = 9.8MRR72 pKa = 11.84KK73 pKa = 8.97KK74 pKa = 9.83MSAEE78 pKa = 3.77GVKK81 pKa = 10.2ILDD84 pKa = 3.76EE85 pKa = 4.22LVNVYY90 pKa = 9.83KK91 pKa = 10.78LKK93 pKa = 10.7SSSPGRR99 pKa = 11.84DD100 pKa = 3.29DD101 pKa = 3.72LTLSRR106 pKa = 11.84VAAAFAAWTCQATEE120 pKa = 3.77AVEE123 pKa = 4.72NYY125 pKa = 9.86MPVTGAAMDD134 pKa = 4.14EE135 pKa = 4.46VSPQYY140 pKa = 9.91PRR142 pKa = 11.84AMMHH146 pKa = 6.66PSFAGLIDD154 pKa = 4.19PQLKK158 pKa = 9.47PDD160 pKa = 3.45QRR162 pKa = 11.84EE163 pKa = 4.23LVVKK167 pKa = 10.44AHH169 pKa = 6.3SLFLIRR175 pKa = 11.84FSRR178 pKa = 11.84VINVALRR185 pKa = 11.84GRR187 pKa = 11.84PKK189 pKa = 10.69SEE191 pKa = 3.75VEE193 pKa = 3.99MSFSQPMNAAINSNFLTSDD212 pKa = 3.02NRR214 pKa = 11.84RR215 pKa = 11.84EE216 pKa = 3.94MLKK219 pKa = 10.71KK220 pKa = 10.65LGIVDD225 pKa = 3.72KK226 pKa = 11.21NGDD229 pKa = 3.72VVPSVIAAARR239 pKa = 11.84AYY241 pKa = 10.43DD242 pKa = 3.73KK243 pKa = 11.01EE244 pKa = 4.17II245 pKa = 4.68
MM1 pKa = 7.36SFSKK5 pKa = 10.44IALEE9 pKa = 4.16FGGEE13 pKa = 4.23SVDD16 pKa = 3.57VDD18 pKa = 4.95EE19 pKa = 4.6ISKK22 pKa = 9.62WVSEE26 pKa = 4.19FAYY29 pKa = 10.11QGYY32 pKa = 9.44DD33 pKa = 2.98ANRR36 pKa = 11.84VIEE39 pKa = 5.45LIQQYY44 pKa = 8.75GRR46 pKa = 11.84KK47 pKa = 9.01RR48 pKa = 11.84DD49 pKa = 2.99WKK51 pKa = 9.14MDD53 pKa = 3.64VKK55 pKa = 11.06RR56 pKa = 11.84MIVLALTRR64 pKa = 11.84GNKK67 pKa = 7.63PDD69 pKa = 3.45KK70 pKa = 9.8MRR72 pKa = 11.84KK73 pKa = 8.97KK74 pKa = 9.83MSAEE78 pKa = 3.77GVKK81 pKa = 10.2ILDD84 pKa = 3.76EE85 pKa = 4.22LVNVYY90 pKa = 9.83KK91 pKa = 10.78LKK93 pKa = 10.7SSSPGRR99 pKa = 11.84DD100 pKa = 3.29DD101 pKa = 3.72LTLSRR106 pKa = 11.84VAAAFAAWTCQATEE120 pKa = 3.77AVEE123 pKa = 4.72NYY125 pKa = 9.86MPVTGAAMDD134 pKa = 4.14EE135 pKa = 4.46VSPQYY140 pKa = 9.91PRR142 pKa = 11.84AMMHH146 pKa = 6.66PSFAGLIDD154 pKa = 4.19PQLKK158 pKa = 9.47PDD160 pKa = 3.45QRR162 pKa = 11.84EE163 pKa = 4.23LVVKK167 pKa = 10.44AHH169 pKa = 6.3SLFLIRR175 pKa = 11.84FSRR178 pKa = 11.84VINVALRR185 pKa = 11.84GRR187 pKa = 11.84PKK189 pKa = 10.69SEE191 pKa = 3.75VEE193 pKa = 3.99MSFSQPMNAAINSNFLTSDD212 pKa = 3.02NRR214 pKa = 11.84RR215 pKa = 11.84EE216 pKa = 3.94MLKK219 pKa = 10.71KK220 pKa = 10.65LGIVDD225 pKa = 3.72KK226 pKa = 11.21NGDD229 pKa = 3.72VVPSVIAAARR239 pKa = 11.84AYY241 pKa = 10.43DD242 pKa = 3.73KK243 pKa = 11.01EE244 pKa = 4.17II245 pKa = 4.68
Molecular weight: 27.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3949 |
245 |
2096 |
987.3 |
111.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.166 ± 0.838 | 2.836 ± 0.758 |
5.9 ± 0.468 | 6.812 ± 0.132 |
4.482 ± 0.469 | 5.9 ± 0.533 |
2.431 ± 0.264 | 7.116 ± 0.566 |
6.761 ± 0.258 | 8.483 ± 0.379 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.343 ± 0.547 | 4.204 ± 0.299 |
3.419 ± 0.551 | 3.191 ± 0.331 |
5.546 ± 0.357 | 9.445 ± 0.598 |
4.685 ± 0.388 | 6.077 ± 0.476 |
1.19 ± 0.043 | 3.013 ± 0.28 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
