
Ruminococcaceae bacterium YRB3002
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; unclassified Oscillospiraceae
Average proteome isoelectric point is 5.69
Get precalculated fractions of proteins
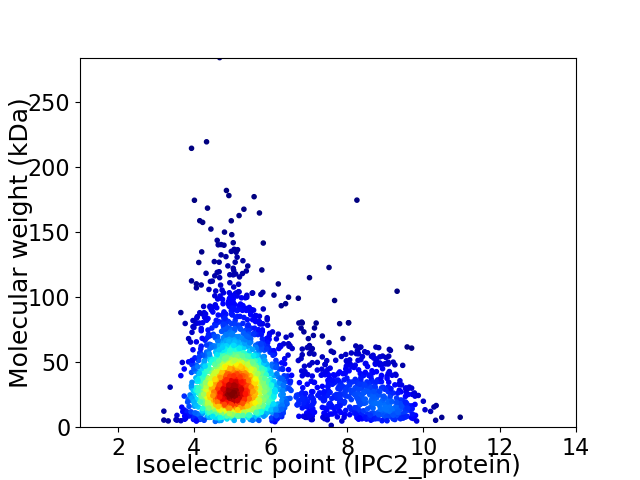
Virtual 2D-PAGE plot for 2644 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
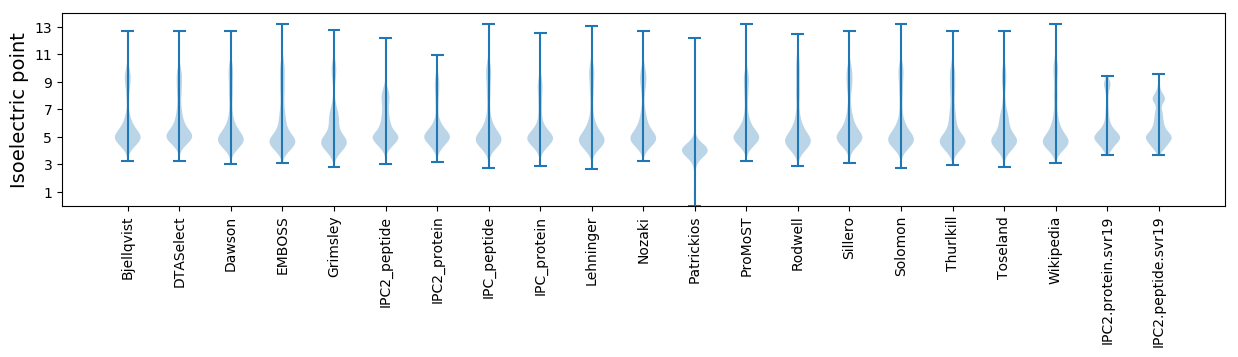
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G4P2K9|A0A1G4P2K9_9FIRM Selenocysteine lyase/Cysteine desulfurase OS=Ruminococcaceae bacterium YRB3002 OX=1520819 GN=SAMN02910456_00060 PE=3 SV=1
MM1 pKa = 7.48INSKK5 pKa = 10.51KK6 pKa = 10.1ILAVVLAGTLALSCAACTSINKK28 pKa = 9.64EE29 pKa = 4.06DD30 pKa = 3.99VIDD33 pKa = 4.1AAEE36 pKa = 4.16SFAKK40 pKa = 10.32AVAALDD46 pKa = 3.79GKK48 pKa = 11.36KK49 pKa = 9.02MLKK52 pKa = 10.27QIDD55 pKa = 4.36DD56 pKa = 3.93IKK58 pKa = 11.11DD59 pKa = 3.42SKK61 pKa = 11.47ADD63 pKa = 3.54QIKK66 pKa = 10.81NKK68 pKa = 10.51LSMSDD73 pKa = 3.98LDD75 pKa = 3.94PDD77 pKa = 3.42EE78 pKa = 5.41AAVKK82 pKa = 10.23KK83 pKa = 10.72AIADD87 pKa = 4.2TITYY91 pKa = 9.51EE92 pKa = 3.82VDD94 pKa = 3.22EE95 pKa = 5.77DD96 pKa = 4.14SVQAGKK102 pKa = 11.04SNDD105 pKa = 3.33EE106 pKa = 4.0ATVDD110 pKa = 3.75VVFTMVDD117 pKa = 3.44YY118 pKa = 11.51AKK120 pKa = 10.9VLTDD124 pKa = 6.41DD125 pKa = 3.79ITTKK129 pKa = 11.29DD130 pKa = 4.08DD131 pKa = 3.99MIDD134 pKa = 3.78AIKK137 pKa = 10.47DD138 pKa = 3.53SKK140 pKa = 9.22KK141 pKa = 7.75TKK143 pKa = 9.13EE144 pKa = 3.99YY145 pKa = 10.26EE146 pKa = 4.1VTFEE150 pKa = 4.9LVADD154 pKa = 4.72DD155 pKa = 4.83GDD157 pKa = 3.79WLIAEE162 pKa = 4.55DD163 pKa = 3.96TLGEE167 pKa = 4.4LGSLYY172 pKa = 10.96DD173 pKa = 3.74FLDD176 pKa = 3.92YY177 pKa = 10.92EE178 pKa = 4.38LSIGGASGNVIDD190 pKa = 5.61MIDD193 pKa = 3.46HH194 pKa = 6.28TSWWMSGVDD203 pKa = 4.41DD204 pKa = 5.01NYY206 pKa = 11.59TNATYY211 pKa = 10.34IEE213 pKa = 4.13FDD215 pKa = 3.21VWFTDD220 pKa = 3.42NPNVSVYY227 pKa = 8.63YY228 pKa = 10.16TVDD231 pKa = 3.13KK232 pKa = 11.17DD233 pKa = 3.58GSQVYY238 pKa = 9.87QSDD241 pKa = 3.84VQTFSGSYY249 pKa = 10.23FEE251 pKa = 5.81AKK253 pKa = 10.68YY254 pKa = 10.76SDD256 pKa = 3.67EE257 pKa = 5.2QNAQMNNGYY266 pKa = 9.29IAAGTYY272 pKa = 9.96KK273 pKa = 10.15IVVFTADD280 pKa = 3.12GTQIADD286 pKa = 3.23GSTTVTVDD294 pKa = 2.62ATTPTTTTSGSDD306 pKa = 3.04TSTEE310 pKa = 4.46FYY312 pKa = 10.05TIHH315 pKa = 7.32DD316 pKa = 4.19GSFADD321 pKa = 3.79IKK323 pKa = 10.94EE324 pKa = 4.2IGWWDD329 pKa = 3.59YY330 pKa = 11.84GVDD333 pKa = 4.39DD334 pKa = 6.29DD335 pKa = 6.58GDD337 pKa = 3.96GEE339 pKa = 4.3NDD341 pKa = 3.82HH342 pKa = 7.45GSMAQDD348 pKa = 3.19GVYY351 pKa = 10.74CIDD354 pKa = 3.48TEE356 pKa = 4.35IIAFSIEE363 pKa = 4.12LNKK366 pKa = 10.55EE367 pKa = 3.45ADD369 pKa = 4.34DD370 pKa = 3.94IYY372 pKa = 10.63YY373 pKa = 10.45AYY375 pKa = 10.9YY376 pKa = 10.1FLPGEE381 pKa = 4.86DD382 pKa = 3.26VDD384 pKa = 5.45ASNIDD389 pKa = 3.49YY390 pKa = 11.03SNPTFADD397 pKa = 4.16TISATSYY404 pKa = 11.31GNGTIYY410 pKa = 11.07YY411 pKa = 9.44NIDD414 pKa = 3.35YY415 pKa = 10.12EE416 pKa = 4.24PDD418 pKa = 3.17KK419 pKa = 10.95MEE421 pKa = 3.9VGTYY425 pKa = 10.46VLVIAKK431 pKa = 9.98DD432 pKa = 3.44AGSVSNPYY440 pKa = 8.15VTAVCSVISSPSSEE454 pKa = 4.55FINN457 pKa = 4.03
MM1 pKa = 7.48INSKK5 pKa = 10.51KK6 pKa = 10.1ILAVVLAGTLALSCAACTSINKK28 pKa = 9.64EE29 pKa = 4.06DD30 pKa = 3.99VIDD33 pKa = 4.1AAEE36 pKa = 4.16SFAKK40 pKa = 10.32AVAALDD46 pKa = 3.79GKK48 pKa = 11.36KK49 pKa = 9.02MLKK52 pKa = 10.27QIDD55 pKa = 4.36DD56 pKa = 3.93IKK58 pKa = 11.11DD59 pKa = 3.42SKK61 pKa = 11.47ADD63 pKa = 3.54QIKK66 pKa = 10.81NKK68 pKa = 10.51LSMSDD73 pKa = 3.98LDD75 pKa = 3.94PDD77 pKa = 3.42EE78 pKa = 5.41AAVKK82 pKa = 10.23KK83 pKa = 10.72AIADD87 pKa = 4.2TITYY91 pKa = 9.51EE92 pKa = 3.82VDD94 pKa = 3.22EE95 pKa = 5.77DD96 pKa = 4.14SVQAGKK102 pKa = 11.04SNDD105 pKa = 3.33EE106 pKa = 4.0ATVDD110 pKa = 3.75VVFTMVDD117 pKa = 3.44YY118 pKa = 11.51AKK120 pKa = 10.9VLTDD124 pKa = 6.41DD125 pKa = 3.79ITTKK129 pKa = 11.29DD130 pKa = 4.08DD131 pKa = 3.99MIDD134 pKa = 3.78AIKK137 pKa = 10.47DD138 pKa = 3.53SKK140 pKa = 9.22KK141 pKa = 7.75TKK143 pKa = 9.13EE144 pKa = 3.99YY145 pKa = 10.26EE146 pKa = 4.1VTFEE150 pKa = 4.9LVADD154 pKa = 4.72DD155 pKa = 4.83GDD157 pKa = 3.79WLIAEE162 pKa = 4.55DD163 pKa = 3.96TLGEE167 pKa = 4.4LGSLYY172 pKa = 10.96DD173 pKa = 3.74FLDD176 pKa = 3.92YY177 pKa = 10.92EE178 pKa = 4.38LSIGGASGNVIDD190 pKa = 5.61MIDD193 pKa = 3.46HH194 pKa = 6.28TSWWMSGVDD203 pKa = 4.41DD204 pKa = 5.01NYY206 pKa = 11.59TNATYY211 pKa = 10.34IEE213 pKa = 4.13FDD215 pKa = 3.21VWFTDD220 pKa = 3.42NPNVSVYY227 pKa = 8.63YY228 pKa = 10.16TVDD231 pKa = 3.13KK232 pKa = 11.17DD233 pKa = 3.58GSQVYY238 pKa = 9.87QSDD241 pKa = 3.84VQTFSGSYY249 pKa = 10.23FEE251 pKa = 5.81AKK253 pKa = 10.68YY254 pKa = 10.76SDD256 pKa = 3.67EE257 pKa = 5.2QNAQMNNGYY266 pKa = 9.29IAAGTYY272 pKa = 9.96KK273 pKa = 10.15IVVFTADD280 pKa = 3.12GTQIADD286 pKa = 3.23GSTTVTVDD294 pKa = 2.62ATTPTTTTSGSDD306 pKa = 3.04TSTEE310 pKa = 4.46FYY312 pKa = 10.05TIHH315 pKa = 7.32DD316 pKa = 4.19GSFADD321 pKa = 3.79IKK323 pKa = 10.94EE324 pKa = 4.2IGWWDD329 pKa = 3.59YY330 pKa = 11.84GVDD333 pKa = 4.39DD334 pKa = 6.29DD335 pKa = 6.58GDD337 pKa = 3.96GEE339 pKa = 4.3NDD341 pKa = 3.82HH342 pKa = 7.45GSMAQDD348 pKa = 3.19GVYY351 pKa = 10.74CIDD354 pKa = 3.48TEE356 pKa = 4.35IIAFSIEE363 pKa = 4.12LNKK366 pKa = 10.55EE367 pKa = 3.45ADD369 pKa = 4.34DD370 pKa = 3.94IYY372 pKa = 10.63YY373 pKa = 10.45AYY375 pKa = 10.9YY376 pKa = 10.1FLPGEE381 pKa = 4.86DD382 pKa = 3.26VDD384 pKa = 5.45ASNIDD389 pKa = 3.49YY390 pKa = 11.03SNPTFADD397 pKa = 4.16TISATSYY404 pKa = 11.31GNGTIYY410 pKa = 11.07YY411 pKa = 9.44NIDD414 pKa = 3.35YY415 pKa = 10.12EE416 pKa = 4.24PDD418 pKa = 3.17KK419 pKa = 10.95MEE421 pKa = 3.9VGTYY425 pKa = 10.46VLVIAKK431 pKa = 9.98DD432 pKa = 3.44AGSVSNPYY440 pKa = 8.15VTAVCSVISSPSSEE454 pKa = 4.55FINN457 pKa = 4.03
Molecular weight: 49.86 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G4Q3X9|A0A1G4Q3X9_9FIRM Uncharacterized protein OS=Ruminococcaceae bacterium YRB3002 OX=1520819 GN=SAMN02910456_00854 PE=4 SV=1
MM1 pKa = 7.76FITMPFCRR9 pKa = 11.84NAIGNKK15 pKa = 7.12IWRR18 pKa = 11.84TKK20 pKa = 9.0AMKK23 pKa = 8.36MTFQPKK29 pKa = 8.95KK30 pKa = 7.58RR31 pKa = 11.84QRR33 pKa = 11.84SKK35 pKa = 9.17VHH37 pKa = 5.88GFRR40 pKa = 11.84ARR42 pKa = 11.84MRR44 pKa = 11.84TAGGRR49 pKa = 11.84KK50 pKa = 8.31VLARR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84LKK58 pKa = 10.68GRR60 pKa = 11.84KK61 pKa = 8.39QLAAA65 pKa = 4.14
MM1 pKa = 7.76FITMPFCRR9 pKa = 11.84NAIGNKK15 pKa = 7.12IWRR18 pKa = 11.84TKK20 pKa = 9.0AMKK23 pKa = 8.36MTFQPKK29 pKa = 8.95KK30 pKa = 7.58RR31 pKa = 11.84QRR33 pKa = 11.84SKK35 pKa = 9.17VHH37 pKa = 5.88GFRR40 pKa = 11.84ARR42 pKa = 11.84MRR44 pKa = 11.84TAGGRR49 pKa = 11.84KK50 pKa = 8.31VLARR54 pKa = 11.84RR55 pKa = 11.84RR56 pKa = 11.84LKK58 pKa = 10.68GRR60 pKa = 11.84KK61 pKa = 8.39QLAAA65 pKa = 4.14
Molecular weight: 7.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
907111 |
10 |
2697 |
343.1 |
38.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.518 ± 0.038 | 1.567 ± 0.019 |
6.752 ± 0.042 | 6.664 ± 0.05 |
4.201 ± 0.032 | 7.263 ± 0.045 |
1.594 ± 0.015 | 7.608 ± 0.037 |
5.694 ± 0.046 | 8.3 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.02 ± 0.024 | 4.33 ± 0.029 |
3.517 ± 0.028 | 2.375 ± 0.022 |
4.769 ± 0.035 | 6.57 ± 0.036 |
5.71 ± 0.047 | 7.25 ± 0.038 |
0.922 ± 0.017 | 4.377 ± 0.037 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
