
Pelagibacter phage HTVC011P
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Autographiviridae; Stopavirus; Pelagibacter virus HTVC011P
Average proteome isoelectric point is 7.19
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 45 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|M1I885|M1I885_9CAUD Uncharacterized protein OS=Pelagibacter phage HTVC011P OX=1283078 PE=4 SV=1
MM1 pKa = 6.7TQYY4 pKa = 11.75AFNTYY9 pKa = 8.45TGNGSTTQYY18 pKa = 10.49SISFSYY24 pKa = 10.51IDD26 pKa = 3.64STHH29 pKa = 5.73VKK31 pKa = 10.12CFLDD35 pKa = 4.26GVEE38 pKa = 4.54TSAFTISSSTVTFNTAPTNSSVIRR62 pKa = 11.84IEE64 pKa = 3.81RR65 pKa = 11.84QTPVDD70 pKa = 3.48SRR72 pKa = 11.84LVDD75 pKa = 4.21FQDD78 pKa = 3.99GSVLTEE84 pKa = 4.27AEE86 pKa = 4.39LDD88 pKa = 3.35MSANQNFYY96 pKa = 10.95AVQEE100 pKa = 4.28ITDD103 pKa = 3.93DD104 pKa = 3.5QSNNLGLTTADD115 pKa = 4.08VYY117 pKa = 11.31DD118 pKa = 4.45ANNKK122 pKa = 10.0RR123 pKa = 11.84IINVADD129 pKa = 3.7PVDD132 pKa = 3.57NTDD135 pKa = 3.33AVNKK139 pKa = 9.92QFISTNLPNITTVAGISADD158 pKa = 3.48VTTVAGISADD168 pKa = 3.58VTAVANDD175 pKa = 3.36ATDD178 pKa = 3.12IGVVSTNIASVNTVATNIADD198 pKa = 4.11VIKK201 pKa = 11.04VADD204 pKa = 4.79DD205 pKa = 3.71LNEE208 pKa = 4.8AISEE212 pKa = 4.4VEE214 pKa = 4.24TVANDD219 pKa = 3.58LNEE222 pKa = 4.22ATSEE226 pKa = 4.16IEE228 pKa = 4.1VVANNIVNVNTVGTINADD246 pKa = 3.27VTTVANNEE254 pKa = 3.83TDD256 pKa = 3.26IQTLADD262 pKa = 4.24LEE264 pKa = 5.04DD265 pKa = 4.01GTVTTNGLSTLAGLNTEE282 pKa = 3.89IQGVYY287 pKa = 10.52NIRR290 pKa = 11.84TNVTNVDD297 pKa = 3.54TNATNVNLVAGQISPTNNVSAVSAVATEE325 pKa = 3.92IGTLGALGTEE335 pKa = 3.91ITNLNNIRR343 pKa = 11.84TDD345 pKa = 2.9ISGVNTISAEE355 pKa = 4.07VTAVNNNSTNINTVAGLDD373 pKa = 3.74TEE375 pKa = 4.45ITALGASGTVASINTVASNLASVNNFGEE403 pKa = 4.61VYY405 pKa = 10.0RR406 pKa = 11.84ISATAPTTSLDD417 pKa = 4.65DD418 pKa = 3.64GDD420 pKa = 3.69MWFDD424 pKa = 3.38TTADD428 pKa = 3.93KK429 pKa = 11.3LKK431 pKa = 9.82IWNGSSFDD439 pKa = 4.06LAGSSINGTSARR451 pKa = 11.84FKK453 pKa = 10.58YY454 pKa = 10.27VATAGQTTFTGADD467 pKa = 3.31ANGEE471 pKa = 4.13TLAYY475 pKa = 7.87DD476 pKa = 3.59TGYY479 pKa = 10.53IDD481 pKa = 5.3AYY483 pKa = 11.15LNGVHH488 pKa = 7.25LDD490 pKa = 3.62PSDD493 pKa = 3.59YY494 pKa = 10.66TATDD498 pKa = 3.45GSNFVLSSGATVSDD512 pKa = 3.4EE513 pKa = 4.28LYY515 pKa = 10.1IVAFGTFSVANIDD528 pKa = 3.74ASNISAGTINEE539 pKa = 3.83ARR541 pKa = 11.84LPTITNAKK549 pKa = 9.29LDD551 pKa = 3.8NSSITINGSAVNLGGSVTVGEE572 pKa = 4.52TKK574 pKa = 9.18PTISSISPDD583 pKa = 3.97TITNAQTSITITGSNFVSVPQVEE606 pKa = 4.51FLNPSTGIWYY616 pKa = 9.82VADD619 pKa = 3.22TVTYY623 pKa = 10.71NNSTSLTVNATLGVDD638 pKa = 3.47GTIQNKK644 pKa = 8.43NRR646 pKa = 11.84EE647 pKa = 4.3PRR649 pKa = 11.84WFSNIIWNNTYY660 pKa = 10.12SFRR663 pKa = 11.84CTNMEE668 pKa = 4.22DD669 pKa = 3.79CCWRR673 pKa = 11.84YY674 pKa = 10.01
MM1 pKa = 6.7TQYY4 pKa = 11.75AFNTYY9 pKa = 8.45TGNGSTTQYY18 pKa = 10.49SISFSYY24 pKa = 10.51IDD26 pKa = 3.64STHH29 pKa = 5.73VKK31 pKa = 10.12CFLDD35 pKa = 4.26GVEE38 pKa = 4.54TSAFTISSSTVTFNTAPTNSSVIRR62 pKa = 11.84IEE64 pKa = 3.81RR65 pKa = 11.84QTPVDD70 pKa = 3.48SRR72 pKa = 11.84LVDD75 pKa = 4.21FQDD78 pKa = 3.99GSVLTEE84 pKa = 4.27AEE86 pKa = 4.39LDD88 pKa = 3.35MSANQNFYY96 pKa = 10.95AVQEE100 pKa = 4.28ITDD103 pKa = 3.93DD104 pKa = 3.5QSNNLGLTTADD115 pKa = 4.08VYY117 pKa = 11.31DD118 pKa = 4.45ANNKK122 pKa = 10.0RR123 pKa = 11.84IINVADD129 pKa = 3.7PVDD132 pKa = 3.57NTDD135 pKa = 3.33AVNKK139 pKa = 9.92QFISTNLPNITTVAGISADD158 pKa = 3.48VTTVAGISADD168 pKa = 3.58VTAVANDD175 pKa = 3.36ATDD178 pKa = 3.12IGVVSTNIASVNTVATNIADD198 pKa = 4.11VIKK201 pKa = 11.04VADD204 pKa = 4.79DD205 pKa = 3.71LNEE208 pKa = 4.8AISEE212 pKa = 4.4VEE214 pKa = 4.24TVANDD219 pKa = 3.58LNEE222 pKa = 4.22ATSEE226 pKa = 4.16IEE228 pKa = 4.1VVANNIVNVNTVGTINADD246 pKa = 3.27VTTVANNEE254 pKa = 3.83TDD256 pKa = 3.26IQTLADD262 pKa = 4.24LEE264 pKa = 5.04DD265 pKa = 4.01GTVTTNGLSTLAGLNTEE282 pKa = 3.89IQGVYY287 pKa = 10.52NIRR290 pKa = 11.84TNVTNVDD297 pKa = 3.54TNATNVNLVAGQISPTNNVSAVSAVATEE325 pKa = 3.92IGTLGALGTEE335 pKa = 3.91ITNLNNIRR343 pKa = 11.84TDD345 pKa = 2.9ISGVNTISAEE355 pKa = 4.07VTAVNNNSTNINTVAGLDD373 pKa = 3.74TEE375 pKa = 4.45ITALGASGTVASINTVASNLASVNNFGEE403 pKa = 4.61VYY405 pKa = 10.0RR406 pKa = 11.84ISATAPTTSLDD417 pKa = 4.65DD418 pKa = 3.64GDD420 pKa = 3.69MWFDD424 pKa = 3.38TTADD428 pKa = 3.93KK429 pKa = 11.3LKK431 pKa = 9.82IWNGSSFDD439 pKa = 4.06LAGSSINGTSARR451 pKa = 11.84FKK453 pKa = 10.58YY454 pKa = 10.27VATAGQTTFTGADD467 pKa = 3.31ANGEE471 pKa = 4.13TLAYY475 pKa = 7.87DD476 pKa = 3.59TGYY479 pKa = 10.53IDD481 pKa = 5.3AYY483 pKa = 11.15LNGVHH488 pKa = 7.25LDD490 pKa = 3.62PSDD493 pKa = 3.59YY494 pKa = 10.66TATDD498 pKa = 3.45GSNFVLSSGATVSDD512 pKa = 3.4EE513 pKa = 4.28LYY515 pKa = 10.1IVAFGTFSVANIDD528 pKa = 3.74ASNISAGTINEE539 pKa = 3.83ARR541 pKa = 11.84LPTITNAKK549 pKa = 9.29LDD551 pKa = 3.8NSSITINGSAVNLGGSVTVGEE572 pKa = 4.52TKK574 pKa = 9.18PTISSISPDD583 pKa = 3.97TITNAQTSITITGSNFVSVPQVEE606 pKa = 4.51FLNPSTGIWYY616 pKa = 9.82VADD619 pKa = 3.22TVTYY623 pKa = 10.71NNSTSLTVNATLGVDD638 pKa = 3.47GTIQNKK644 pKa = 8.43NRR646 pKa = 11.84EE647 pKa = 4.3PRR649 pKa = 11.84WFSNIIWNNTYY660 pKa = 10.12SFRR663 pKa = 11.84CTNMEE668 pKa = 4.22DD669 pKa = 3.79CCWRR673 pKa = 11.84YY674 pKa = 10.01
Molecular weight: 70.79 kDa
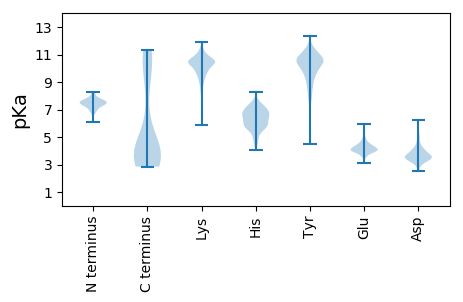
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M1HM84|M1HM84_9CAUD Uncharacterized protein OS=Pelagibacter phage HTVC011P OX=1283078 PE=4 SV=1
MM1 pKa = 7.49SNATTKK7 pKa = 10.55QKK9 pKa = 11.26APTEE13 pKa = 4.13ASTDD17 pKa = 3.23ASRR20 pKa = 11.84DD21 pKa = 3.4TTVASNYY28 pKa = 7.92SRR30 pKa = 11.84KK31 pKa = 9.42RR32 pKa = 11.84VGRR35 pKa = 11.84GSLRR39 pKa = 11.84IPLSGGSGLNFPTSS53 pKa = 3.19
MM1 pKa = 7.49SNATTKK7 pKa = 10.55QKK9 pKa = 11.26APTEE13 pKa = 4.13ASTDD17 pKa = 3.23ASRR20 pKa = 11.84DD21 pKa = 3.4TTVASNYY28 pKa = 7.92SRR30 pKa = 11.84KK31 pKa = 9.42RR32 pKa = 11.84VGRR35 pKa = 11.84GSLRR39 pKa = 11.84IPLSGGSGLNFPTSS53 pKa = 3.19
Molecular weight: 5.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12316 |
41 |
1230 |
273.7 |
30.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.504 ± 0.3 | 0.796 ± 0.165 |
5.83 ± 0.242 | 6.025 ± 0.417 |
4.117 ± 0.228 | 6.398 ± 0.37 |
1.575 ± 0.201 | 6.983 ± 0.291 |
8.469 ± 0.652 | 8.071 ± 0.383 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.135 ± 0.187 | 6.699 ± 0.375 |
2.939 ± 0.162 | 3.979 ± 0.309 |
3.784 ± 0.262 | 7.616 ± 0.57 |
7.584 ± 0.65 | 5.781 ± 0.373 |
1.056 ± 0.139 | 3.662 ± 0.196 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
