
Hubei sobemo-like virus 35
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.13
Get precalculated fractions of proteins
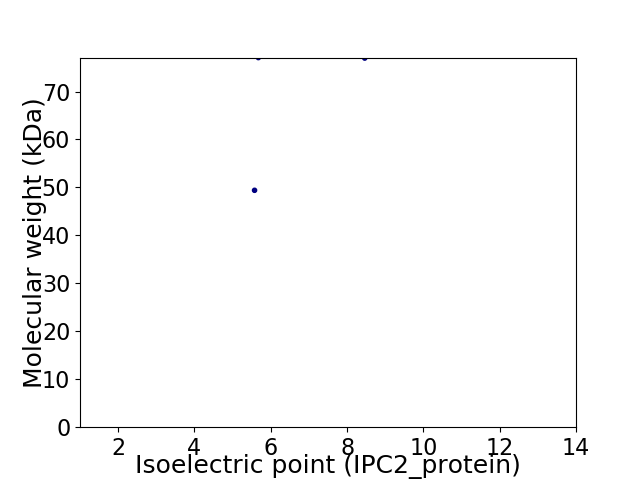
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEI2|A0A1L3KEI2_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 35 OX=1923222 PE=4 SV=1
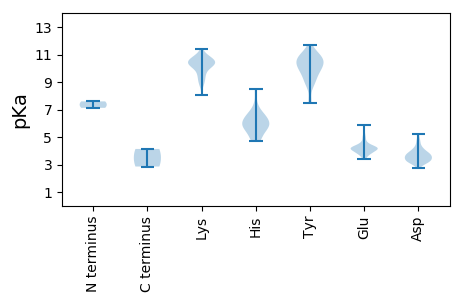
MM1 pKa = 7.12PQRR4 pKa = 11.84YY5 pKa = 8.84DD6 pKa = 2.73WGQDD10 pKa = 3.09VSYY13 pKa = 11.15LEE15 pKa = 4.42EE16 pKa = 4.16LSKK19 pKa = 10.82EE20 pKa = 4.14YY21 pKa = 10.3SWPSFGHH28 pKa = 4.69TAEE31 pKa = 3.83IHH33 pKa = 4.95SLRR36 pKa = 11.84WHH38 pKa = 6.44LPLRR42 pKa = 11.84DD43 pKa = 3.64RR44 pKa = 11.84VEE46 pKa = 3.91QTPPQDD52 pKa = 3.2KK53 pKa = 10.27EE54 pKa = 4.35AILGEE59 pKa = 4.17TTKK62 pKa = 11.06YY63 pKa = 9.02VTDD66 pKa = 3.57AMAFIPEE73 pKa = 4.63DD74 pKa = 3.56FMQYY78 pKa = 7.54SHH80 pKa = 6.12YY81 pKa = 10.5VRR83 pKa = 11.84VVKK86 pKa = 10.22EE87 pKa = 4.29LEE89 pKa = 4.07WDD91 pKa = 3.61SSPGFPYY98 pKa = 9.77MYY100 pKa = 9.44EE101 pKa = 4.15YY102 pKa = 10.53PNNRR106 pKa = 11.84AFFRR110 pKa = 11.84VKK112 pKa = 10.58DD113 pKa = 3.88GLPDD117 pKa = 3.73PEE119 pKa = 5.04RR120 pKa = 11.84VDD122 pKa = 4.04ACWAMVQEE130 pKa = 4.28RR131 pKa = 11.84LKK133 pKa = 11.27NRR135 pKa = 11.84DD136 pKa = 3.08SDD138 pKa = 4.59PIRR141 pKa = 11.84LFIKK145 pKa = 10.25PEE147 pKa = 3.55PHH149 pKa = 5.44TKK151 pKa = 10.35KK152 pKa = 10.57KK153 pKa = 10.27IEE155 pKa = 4.05EE156 pKa = 3.86KK157 pKa = 10.2RR158 pKa = 11.84FRR160 pKa = 11.84LISSVSVVDD169 pKa = 4.32QILDD173 pKa = 3.28QMIFGFQNQAFLDD186 pKa = 4.18NHH188 pKa = 5.96HH189 pKa = 6.61HH190 pKa = 5.55TPVRR194 pKa = 11.84VGWGWMKK201 pKa = 10.88GGWNSVPKK209 pKa = 10.26SGMVACDD216 pKa = 3.15KK217 pKa = 10.85TGWDD221 pKa = 3.4WTVSAWLVDD230 pKa = 3.87LEE232 pKa = 4.45LEE234 pKa = 4.17LRR236 pKa = 11.84SRR238 pKa = 11.84LIFGTEE244 pKa = 3.38KK245 pKa = 10.98SLWEE249 pKa = 3.98DD250 pKa = 3.53LARR253 pKa = 11.84FRR255 pKa = 11.84YY256 pKa = 9.74HH257 pKa = 6.79EE258 pKa = 4.38LFIKK262 pKa = 10.72NEE264 pKa = 3.74FMTSGGLVFRR274 pKa = 11.84VRR276 pKa = 11.84QPGLMKK282 pKa = 10.34SGCVNTIVSNSLMQLLLHH300 pKa = 5.74VRR302 pKa = 11.84VSIEE306 pKa = 4.12LGEE309 pKa = 4.15QIHH312 pKa = 6.6NIWAMGDD319 pKa = 3.43DD320 pKa = 4.81TIQDD324 pKa = 3.7FMPFLPEE331 pKa = 3.67YY332 pKa = 10.96AEE334 pKa = 4.23LLGRR338 pKa = 11.84YY339 pKa = 9.05CILKK343 pKa = 9.8QVSGDD348 pKa = 3.68SEE350 pKa = 4.26FAGFRR355 pKa = 11.84WDD357 pKa = 3.21GDD359 pKa = 4.24YY360 pKa = 10.93IEE362 pKa = 4.89PLYY365 pKa = 10.08PEE367 pKa = 3.8KK368 pKa = 10.62HH369 pKa = 5.98AFQILHH375 pKa = 5.94VKK377 pKa = 10.26EE378 pKa = 3.8KK379 pKa = 10.29DD380 pKa = 3.22KK381 pKa = 11.44KK382 pKa = 10.72VFSLSYY388 pKa = 11.0NLLYY392 pKa = 10.01WKK394 pKa = 10.52SRR396 pKa = 11.84FLPYY400 pKa = 9.77IQRR403 pKa = 11.84LMPVPDD409 pKa = 3.19IGFTRR414 pKa = 11.84IWDD417 pKa = 3.79GEE419 pKa = 4.13
MM1 pKa = 7.12PQRR4 pKa = 11.84YY5 pKa = 8.84DD6 pKa = 2.73WGQDD10 pKa = 3.09VSYY13 pKa = 11.15LEE15 pKa = 4.42EE16 pKa = 4.16LSKK19 pKa = 10.82EE20 pKa = 4.14YY21 pKa = 10.3SWPSFGHH28 pKa = 4.69TAEE31 pKa = 3.83IHH33 pKa = 4.95SLRR36 pKa = 11.84WHH38 pKa = 6.44LPLRR42 pKa = 11.84DD43 pKa = 3.64RR44 pKa = 11.84VEE46 pKa = 3.91QTPPQDD52 pKa = 3.2KK53 pKa = 10.27EE54 pKa = 4.35AILGEE59 pKa = 4.17TTKK62 pKa = 11.06YY63 pKa = 9.02VTDD66 pKa = 3.57AMAFIPEE73 pKa = 4.63DD74 pKa = 3.56FMQYY78 pKa = 7.54SHH80 pKa = 6.12YY81 pKa = 10.5VRR83 pKa = 11.84VVKK86 pKa = 10.22EE87 pKa = 4.29LEE89 pKa = 4.07WDD91 pKa = 3.61SSPGFPYY98 pKa = 9.77MYY100 pKa = 9.44EE101 pKa = 4.15YY102 pKa = 10.53PNNRR106 pKa = 11.84AFFRR110 pKa = 11.84VKK112 pKa = 10.58DD113 pKa = 3.88GLPDD117 pKa = 3.73PEE119 pKa = 5.04RR120 pKa = 11.84VDD122 pKa = 4.04ACWAMVQEE130 pKa = 4.28RR131 pKa = 11.84LKK133 pKa = 11.27NRR135 pKa = 11.84DD136 pKa = 3.08SDD138 pKa = 4.59PIRR141 pKa = 11.84LFIKK145 pKa = 10.25PEE147 pKa = 3.55PHH149 pKa = 5.44TKK151 pKa = 10.35KK152 pKa = 10.57KK153 pKa = 10.27IEE155 pKa = 4.05EE156 pKa = 3.86KK157 pKa = 10.2RR158 pKa = 11.84FRR160 pKa = 11.84LISSVSVVDD169 pKa = 4.32QILDD173 pKa = 3.28QMIFGFQNQAFLDD186 pKa = 4.18NHH188 pKa = 5.96HH189 pKa = 6.61HH190 pKa = 5.55TPVRR194 pKa = 11.84VGWGWMKK201 pKa = 10.88GGWNSVPKK209 pKa = 10.26SGMVACDD216 pKa = 3.15KK217 pKa = 10.85TGWDD221 pKa = 3.4WTVSAWLVDD230 pKa = 3.87LEE232 pKa = 4.45LEE234 pKa = 4.17LRR236 pKa = 11.84SRR238 pKa = 11.84LIFGTEE244 pKa = 3.38KK245 pKa = 10.98SLWEE249 pKa = 3.98DD250 pKa = 3.53LARR253 pKa = 11.84FRR255 pKa = 11.84YY256 pKa = 9.74HH257 pKa = 6.79EE258 pKa = 4.38LFIKK262 pKa = 10.72NEE264 pKa = 3.74FMTSGGLVFRR274 pKa = 11.84VRR276 pKa = 11.84QPGLMKK282 pKa = 10.34SGCVNTIVSNSLMQLLLHH300 pKa = 5.74VRR302 pKa = 11.84VSIEE306 pKa = 4.12LGEE309 pKa = 4.15QIHH312 pKa = 6.6NIWAMGDD319 pKa = 3.43DD320 pKa = 4.81TIQDD324 pKa = 3.7FMPFLPEE331 pKa = 3.67YY332 pKa = 10.96AEE334 pKa = 4.23LLGRR338 pKa = 11.84YY339 pKa = 9.05CILKK343 pKa = 9.8QVSGDD348 pKa = 3.68SEE350 pKa = 4.26FAGFRR355 pKa = 11.84WDD357 pKa = 3.21GDD359 pKa = 4.24YY360 pKa = 10.93IEE362 pKa = 4.89PLYY365 pKa = 10.08PEE367 pKa = 3.8KK368 pKa = 10.62HH369 pKa = 5.98AFQILHH375 pKa = 5.94VKK377 pKa = 10.26EE378 pKa = 3.8KK379 pKa = 10.29DD380 pKa = 3.22KK381 pKa = 11.44KK382 pKa = 10.72VFSLSYY388 pKa = 11.0NLLYY392 pKa = 10.01WKK394 pKa = 10.52SRR396 pKa = 11.84FLPYY400 pKa = 9.77IQRR403 pKa = 11.84LMPVPDD409 pKa = 3.19IGFTRR414 pKa = 11.84IWDD417 pKa = 3.79GEE419 pKa = 4.13
Molecular weight: 49.4 kDa
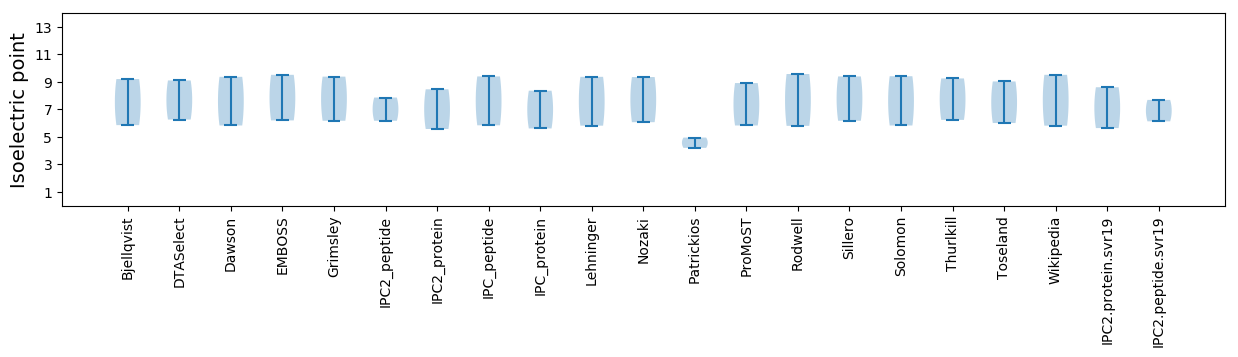
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEI2|A0A1L3KEI2_9VIRU RNA-directed RNA polymerase OS=Hubei sobemo-like virus 35 OX=1923222 PE=4 SV=1
MM1 pKa = 7.59EE2 pKa = 5.88SIKK5 pKa = 10.55QLALEE10 pKa = 4.21TLQNAGQLQLVGCLIILAVVVSNRR34 pKa = 11.84RR35 pKa = 11.84LITFPMVPLALWLILWDD52 pKa = 3.42TRR54 pKa = 11.84EE55 pKa = 4.14RR56 pKa = 11.84IVYY59 pKa = 9.54RR60 pKa = 11.84PVMPDD65 pKa = 3.5FSWVQAWVLMASEE78 pKa = 4.51SFQRR82 pKa = 11.84AFEE85 pKa = 4.03ASKK88 pKa = 10.52AYY90 pKa = 9.1MMGVIGVMVLLRR102 pKa = 11.84VWISLRR108 pKa = 11.84QSLKK112 pKa = 10.32EE113 pKa = 3.55VSLRR117 pKa = 11.84LRR119 pKa = 11.84GVTRR123 pKa = 11.84VQYY126 pKa = 10.03EE127 pKa = 3.83AMRR130 pKa = 11.84EE131 pKa = 3.88GSAFEE136 pKa = 4.18KK137 pKa = 10.93APIPACQVQICEE149 pKa = 4.22AGLFVNSHH157 pKa = 6.65LGYY160 pKa = 10.46GIRR163 pKa = 11.84VEE165 pKa = 4.16NCLVVPRR172 pKa = 11.84HH173 pKa = 5.56VLLAGKK179 pKa = 8.98WPTNKK184 pKa = 10.38LIISVPLRR192 pKa = 11.84PSMVLEE198 pKa = 3.94GRR200 pKa = 11.84MEE202 pKa = 4.06EE203 pKa = 4.2SQKK206 pKa = 10.59FDD208 pKa = 3.21LAYY211 pKa = 10.59FFLGEE216 pKa = 4.2AQWAALGAKK225 pKa = 9.59KK226 pKa = 9.34ATLTASSLQGRR237 pKa = 11.84ASCAGYY243 pKa = 10.74AGLSRR248 pKa = 11.84GDD250 pKa = 3.62LKK252 pKa = 10.54KK253 pKa = 9.41TNSVGQYY260 pKa = 10.37SYY262 pKa = 11.66GGSTKK267 pKa = 10.52AGYY270 pKa = 10.07SGAAYY275 pKa = 10.53VMQGQVAGIHH285 pKa = 6.18IGVDD289 pKa = 3.39CEE291 pKa = 4.67VNVGLSSRR299 pKa = 11.84LIAKK303 pKa = 8.97EE304 pKa = 3.91LEE306 pKa = 4.22YY307 pKa = 10.52LTVTEE312 pKa = 4.0ARR314 pKa = 11.84RR315 pKa = 11.84NHH317 pKa = 5.83GVSGLAAYY325 pKa = 7.47DD326 pKa = 3.71TPVQLVEE333 pKa = 4.58THH335 pKa = 7.36DD336 pKa = 3.16NWQDD340 pKa = 3.24LDD342 pKa = 3.83IEE344 pKa = 4.29EE345 pKa = 4.98LMYY348 pKa = 10.87ASRR351 pKa = 11.84SGDD354 pKa = 3.16EE355 pKa = 5.34RR356 pKa = 11.84ATEE359 pKa = 3.99WLRR362 pKa = 11.84KK363 pKa = 8.68NQHH366 pKa = 4.9KK367 pKa = 10.14HH368 pKa = 5.64KK369 pKa = 9.31KK370 pKa = 9.15HH371 pKa = 4.83PMYY374 pKa = 9.87WEE376 pKa = 4.17SMKK379 pKa = 10.63QALPSMDD386 pKa = 4.73PEE388 pKa = 4.34TLLQVQNAFNKK399 pKa = 10.39AFTFLQEE406 pKa = 4.34EE407 pKa = 4.84TCSCSGLLHH416 pKa = 6.39ACEE419 pKa = 4.27GKK421 pKa = 9.86RR422 pKa = 11.84GKK424 pKa = 10.8VIDD427 pKa = 3.92QRR429 pKa = 11.84PSTSRR434 pKa = 11.84VIGQSDD440 pKa = 4.07VPQEE444 pKa = 4.16AEE446 pKa = 3.83QGLTEE451 pKa = 4.44TPLHH455 pKa = 6.14RR456 pKa = 11.84ANMLKK461 pKa = 10.2RR462 pKa = 11.84IEE464 pKa = 4.15SLEE467 pKa = 4.12EE468 pKa = 3.62NEE470 pKa = 4.62KK471 pKa = 10.64ILVEE475 pKa = 4.45SVSNMDD481 pKa = 4.11EE482 pKa = 4.01NVKK485 pKa = 10.29DD486 pKa = 3.46LRR488 pKa = 11.84GNVTQIDD495 pKa = 3.69DD496 pKa = 5.24DD497 pKa = 3.78IYY499 pKa = 11.11KK500 pKa = 10.06IYY502 pKa = 10.77GRR504 pKa = 11.84IAFLEE509 pKa = 3.88EE510 pKa = 3.46RR511 pKa = 11.84LQNVEE516 pKa = 3.41LWAIDD521 pKa = 3.33RR522 pKa = 11.84GFANLPAALFSEE534 pKa = 5.31DD535 pKa = 3.38SPQVVAQKK543 pKa = 10.68RR544 pKa = 11.84RR545 pKa = 11.84TSKK548 pKa = 10.78NPAIPLVEE556 pKa = 5.04RR557 pKa = 11.84ISALKK562 pKa = 10.15LKK564 pKa = 10.56AYY566 pKa = 10.35GEE568 pKa = 4.37STKK571 pKa = 9.93PRR573 pKa = 11.84KK574 pKa = 9.3IVTPPLIRR582 pKa = 11.84KK583 pKa = 8.79VRR585 pKa = 11.84DD586 pKa = 3.85AIATRR591 pKa = 11.84TQSKK595 pKa = 9.73NVEE598 pKa = 4.1VEE600 pKa = 4.16LEE602 pKa = 4.35HH603 pKa = 8.46ISAAEE608 pKa = 3.78EE609 pKa = 3.86ARR611 pKa = 11.84EE612 pKa = 4.22KK613 pKa = 10.14MSSMTQKK620 pKa = 9.71QQRR623 pKa = 11.84EE624 pKa = 4.17RR625 pKa = 11.84YY626 pKa = 8.82KK627 pKa = 10.6EE628 pKa = 3.81LRR630 pKa = 11.84NKK632 pKa = 10.14QNLTPEE638 pKa = 4.24EE639 pKa = 4.11EE640 pKa = 4.29QFKK643 pKa = 9.55QQHH646 pKa = 5.71LEE648 pKa = 3.83SRR650 pKa = 11.84RR651 pKa = 11.84KK652 pKa = 8.95RR653 pKa = 11.84NARR656 pKa = 11.84RR657 pKa = 11.84RR658 pKa = 11.84TGKK661 pKa = 10.37KK662 pKa = 8.03STGNSRR668 pKa = 11.84STSSSSGKK676 pKa = 9.87NEE678 pKa = 4.08PSTSTQQ684 pKa = 2.85
MM1 pKa = 7.59EE2 pKa = 5.88SIKK5 pKa = 10.55QLALEE10 pKa = 4.21TLQNAGQLQLVGCLIILAVVVSNRR34 pKa = 11.84RR35 pKa = 11.84LITFPMVPLALWLILWDD52 pKa = 3.42TRR54 pKa = 11.84EE55 pKa = 4.14RR56 pKa = 11.84IVYY59 pKa = 9.54RR60 pKa = 11.84PVMPDD65 pKa = 3.5FSWVQAWVLMASEE78 pKa = 4.51SFQRR82 pKa = 11.84AFEE85 pKa = 4.03ASKK88 pKa = 10.52AYY90 pKa = 9.1MMGVIGVMVLLRR102 pKa = 11.84VWISLRR108 pKa = 11.84QSLKK112 pKa = 10.32EE113 pKa = 3.55VSLRR117 pKa = 11.84LRR119 pKa = 11.84GVTRR123 pKa = 11.84VQYY126 pKa = 10.03EE127 pKa = 3.83AMRR130 pKa = 11.84EE131 pKa = 3.88GSAFEE136 pKa = 4.18KK137 pKa = 10.93APIPACQVQICEE149 pKa = 4.22AGLFVNSHH157 pKa = 6.65LGYY160 pKa = 10.46GIRR163 pKa = 11.84VEE165 pKa = 4.16NCLVVPRR172 pKa = 11.84HH173 pKa = 5.56VLLAGKK179 pKa = 8.98WPTNKK184 pKa = 10.38LIISVPLRR192 pKa = 11.84PSMVLEE198 pKa = 3.94GRR200 pKa = 11.84MEE202 pKa = 4.06EE203 pKa = 4.2SQKK206 pKa = 10.59FDD208 pKa = 3.21LAYY211 pKa = 10.59FFLGEE216 pKa = 4.2AQWAALGAKK225 pKa = 9.59KK226 pKa = 9.34ATLTASSLQGRR237 pKa = 11.84ASCAGYY243 pKa = 10.74AGLSRR248 pKa = 11.84GDD250 pKa = 3.62LKK252 pKa = 10.54KK253 pKa = 9.41TNSVGQYY260 pKa = 10.37SYY262 pKa = 11.66GGSTKK267 pKa = 10.52AGYY270 pKa = 10.07SGAAYY275 pKa = 10.53VMQGQVAGIHH285 pKa = 6.18IGVDD289 pKa = 3.39CEE291 pKa = 4.67VNVGLSSRR299 pKa = 11.84LIAKK303 pKa = 8.97EE304 pKa = 3.91LEE306 pKa = 4.22YY307 pKa = 10.52LTVTEE312 pKa = 4.0ARR314 pKa = 11.84RR315 pKa = 11.84NHH317 pKa = 5.83GVSGLAAYY325 pKa = 7.47DD326 pKa = 3.71TPVQLVEE333 pKa = 4.58THH335 pKa = 7.36DD336 pKa = 3.16NWQDD340 pKa = 3.24LDD342 pKa = 3.83IEE344 pKa = 4.29EE345 pKa = 4.98LMYY348 pKa = 10.87ASRR351 pKa = 11.84SGDD354 pKa = 3.16EE355 pKa = 5.34RR356 pKa = 11.84ATEE359 pKa = 3.99WLRR362 pKa = 11.84KK363 pKa = 8.68NQHH366 pKa = 4.9KK367 pKa = 10.14HH368 pKa = 5.64KK369 pKa = 9.31KK370 pKa = 9.15HH371 pKa = 4.83PMYY374 pKa = 9.87WEE376 pKa = 4.17SMKK379 pKa = 10.63QALPSMDD386 pKa = 4.73PEE388 pKa = 4.34TLLQVQNAFNKK399 pKa = 10.39AFTFLQEE406 pKa = 4.34EE407 pKa = 4.84TCSCSGLLHH416 pKa = 6.39ACEE419 pKa = 4.27GKK421 pKa = 9.86RR422 pKa = 11.84GKK424 pKa = 10.8VIDD427 pKa = 3.92QRR429 pKa = 11.84PSTSRR434 pKa = 11.84VIGQSDD440 pKa = 4.07VPQEE444 pKa = 4.16AEE446 pKa = 3.83QGLTEE451 pKa = 4.44TPLHH455 pKa = 6.14RR456 pKa = 11.84ANMLKK461 pKa = 10.2RR462 pKa = 11.84IEE464 pKa = 4.15SLEE467 pKa = 4.12EE468 pKa = 3.62NEE470 pKa = 4.62KK471 pKa = 10.64ILVEE475 pKa = 4.45SVSNMDD481 pKa = 4.11EE482 pKa = 4.01NVKK485 pKa = 10.29DD486 pKa = 3.46LRR488 pKa = 11.84GNVTQIDD495 pKa = 3.69DD496 pKa = 5.24DD497 pKa = 3.78IYY499 pKa = 11.11KK500 pKa = 10.06IYY502 pKa = 10.77GRR504 pKa = 11.84IAFLEE509 pKa = 3.88EE510 pKa = 3.46RR511 pKa = 11.84LQNVEE516 pKa = 3.41LWAIDD521 pKa = 3.33RR522 pKa = 11.84GFANLPAALFSEE534 pKa = 5.31DD535 pKa = 3.38SPQVVAQKK543 pKa = 10.68RR544 pKa = 11.84RR545 pKa = 11.84TSKK548 pKa = 10.78NPAIPLVEE556 pKa = 5.04RR557 pKa = 11.84ISALKK562 pKa = 10.15LKK564 pKa = 10.56AYY566 pKa = 10.35GEE568 pKa = 4.37STKK571 pKa = 9.93PRR573 pKa = 11.84KK574 pKa = 9.3IVTPPLIRR582 pKa = 11.84KK583 pKa = 8.79VRR585 pKa = 11.84DD586 pKa = 3.85AIATRR591 pKa = 11.84TQSKK595 pKa = 9.73NVEE598 pKa = 4.1VEE600 pKa = 4.16LEE602 pKa = 4.35HH603 pKa = 8.46ISAAEE608 pKa = 3.78EE609 pKa = 3.86ARR611 pKa = 11.84EE612 pKa = 4.22KK613 pKa = 10.14MSSMTQKK620 pKa = 9.71QQRR623 pKa = 11.84EE624 pKa = 4.17RR625 pKa = 11.84YY626 pKa = 8.82KK627 pKa = 10.6EE628 pKa = 3.81LRR630 pKa = 11.84NKK632 pKa = 10.14QNLTPEE638 pKa = 4.24EE639 pKa = 4.11EE640 pKa = 4.29QFKK643 pKa = 9.55QQHH646 pKa = 5.71LEE648 pKa = 3.83SRR650 pKa = 11.84RR651 pKa = 11.84KK652 pKa = 8.95RR653 pKa = 11.84NARR656 pKa = 11.84RR657 pKa = 11.84RR658 pKa = 11.84TGKK661 pKa = 10.37KK662 pKa = 8.03STGNSRR668 pKa = 11.84STSSSSGKK676 pKa = 9.87NEE678 pKa = 4.08PSTSTQQ684 pKa = 2.85
Molecular weight: 76.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1103 |
419 |
684 |
551.5 |
63.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.437 ± 1.6 | 1.179 ± 0.125 |
4.442 ± 1.254 | 7.978 ± 0.325 |
3.626 ± 1.177 | 5.984 ± 0.01 |
2.267 ± 0.468 | 4.896 ± 0.199 |
5.984 ± 0.143 | 9.701 ± 0.22 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.992 ± 0.196 | 3.354 ± 0.408 |
4.533 ± 0.669 | 5.168 ± 0.622 |
6.981 ± 0.434 | 7.525 ± 0.605 |
4.261 ± 0.515 | 7.072 ± 0.218 |
2.448 ± 0.768 | 3.173 ± 0.495 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
