
Soybean dwarf virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Tolucaviricetes; Tolivirales; Tombusviridae; Luteovirus
Average proteome isoelectric point is 7.19
Get precalculated fractions of proteins
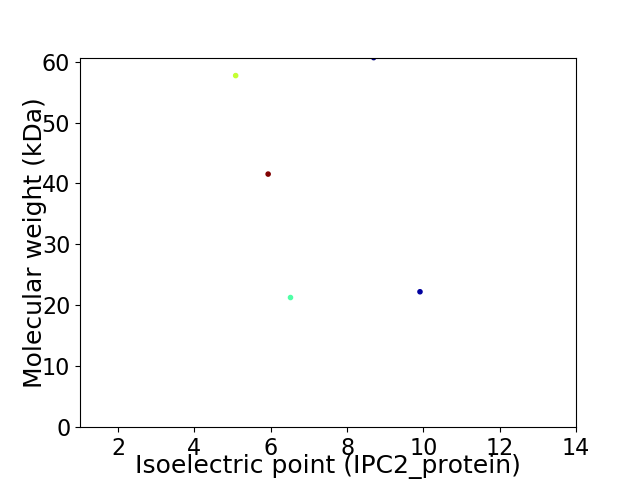
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q91QR2|Q91QR2_9LUTE Movement protein OS=Soybean dwarf virus OX=12049 GN=ORF4 PE=3 SV=1
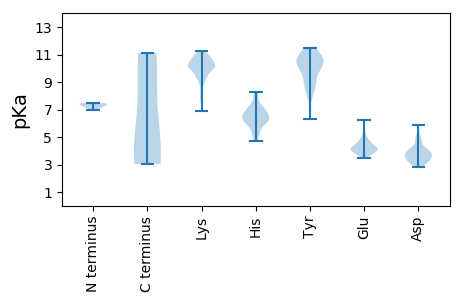
VV1 pKa = 7.46DD2 pKa = 4.87GEE4 pKa = 4.6PGPKK8 pKa = 9.42PGPDD12 pKa = 3.72PAPQPTPTPEE22 pKa = 3.71PTPAKK27 pKa = 9.75HH28 pKa = 6.09EE29 pKa = 4.09RR30 pKa = 11.84FIAYY34 pKa = 7.69TGTLSTLISARR45 pKa = 11.84QSSDD49 pKa = 3.46SISLYY54 pKa = 10.01SIRR57 pKa = 11.84SQRR60 pKa = 11.84IRR62 pKa = 11.84YY63 pKa = 9.1IEE65 pKa = 4.77DD66 pKa = 3.59EE67 pKa = 4.13NSSWTNIDD75 pKa = 3.62AKK77 pKa = 10.16WYY79 pKa = 8.63SQNSVEE85 pKa = 4.73AIPMFVYY92 pKa = 9.48PVPEE96 pKa = 4.3GTWSIEE102 pKa = 3.88ISCEE106 pKa = 3.95GYY108 pKa = 9.81QAASSTSDD116 pKa = 3.17PHH118 pKa = 7.88RR119 pKa = 11.84GKK121 pKa = 10.64CDD123 pKa = 2.94GMIAYY128 pKa = 9.88DD129 pKa = 4.3DD130 pKa = 4.93DD131 pKa = 4.88SSKK134 pKa = 10.55VWNVGQQNNVTITNNKK150 pKa = 9.17ADD152 pKa = 4.07NDD154 pKa = 4.0WKK156 pKa = 11.0YY157 pKa = 11.15GHH159 pKa = 7.75PDD161 pKa = 3.01LTINGDD167 pKa = 3.54RR168 pKa = 11.84FDD170 pKa = 3.77QNQVVEE176 pKa = 4.13KK177 pKa = 10.71DD178 pKa = 3.98GIISFHH184 pKa = 6.69LVTTGPNASFFLVAPAVKK202 pKa = 8.86KK203 pKa = 6.9TAKK206 pKa = 10.58YY207 pKa = 9.35NFCVSYY213 pKa = 11.32GDD215 pKa = 3.19WTDD218 pKa = 3.56RR219 pKa = 11.84DD220 pKa = 3.87MEE222 pKa = 4.49FGMVSVVLDD231 pKa = 3.47EE232 pKa = 4.66HH233 pKa = 8.3LEE235 pKa = 4.22GARR238 pKa = 11.84SSQYY242 pKa = 9.71VRR244 pKa = 11.84KK245 pKa = 7.62TPRR248 pKa = 11.84SGHH251 pKa = 5.3VGVNRR256 pKa = 11.84SHH258 pKa = 7.25RR259 pKa = 11.84LQDD262 pKa = 3.19NFVPTEE268 pKa = 3.97YY269 pKa = 11.14VSDD272 pKa = 3.89EE273 pKa = 4.39DD274 pKa = 4.06SSSNSSIVSNRR285 pKa = 11.84PSTPDD290 pKa = 3.17NDD292 pKa = 3.69SDD294 pKa = 4.09AKK296 pKa = 10.53FAEE299 pKa = 4.34SMKK302 pKa = 10.83GKK304 pKa = 10.29LPSQTKK310 pKa = 10.17LPPKK314 pKa = 10.43GFLSQLSTKK323 pKa = 8.39EE324 pKa = 3.74KK325 pKa = 11.0KK326 pKa = 10.13EE327 pKa = 3.72ISNSKK332 pKa = 8.98PSNVEE337 pKa = 3.79GLVGPLVAAYY347 pKa = 8.65GYY349 pKa = 9.02PSQTGVHH356 pKa = 6.36DD357 pKa = 4.11AARR360 pKa = 11.84EE361 pKa = 3.71ILQAKK366 pKa = 8.14EE367 pKa = 3.51AAEE370 pKa = 3.85NLAEE374 pKa = 4.64LEE376 pKa = 4.14RR377 pKa = 11.84DD378 pKa = 3.2LKK380 pKa = 10.8EE381 pKa = 4.13INKK384 pKa = 9.88LEE386 pKa = 4.37PPDD389 pKa = 4.53VIVQEE394 pKa = 4.66EE395 pKa = 4.43IPDD398 pKa = 3.87FVPPSEE404 pKa = 5.26KK405 pKa = 9.61ILKK408 pKa = 9.96EE409 pKa = 4.11DD410 pKa = 4.1DD411 pKa = 3.75PDD413 pKa = 3.72YY414 pKa = 11.5VPPIWHH420 pKa = 6.66NADD423 pKa = 3.08QAVLVSSYY431 pKa = 10.73EE432 pKa = 3.88PPDD435 pKa = 3.33WSRR438 pKa = 11.84PAYY441 pKa = 10.0EE442 pKa = 5.22SGDD445 pKa = 3.8PPKK448 pKa = 10.29KK449 pKa = 9.24TGTLKK454 pKa = 10.1GTLSKK459 pKa = 11.03LGGSLRR465 pKa = 11.84SGEE468 pKa = 4.1SSLRR472 pKa = 11.84GSLRR476 pKa = 11.84KK477 pKa = 7.81TQDD480 pKa = 2.88QTDD483 pKa = 4.12LDD485 pKa = 4.08NKK487 pKa = 10.25LSKK490 pKa = 10.77LSVIQRR496 pKa = 11.84SRR498 pKa = 11.84YY499 pKa = 8.05QRR501 pKa = 11.84ILNNLGKK508 pKa = 9.24MRR510 pKa = 11.84ARR512 pKa = 11.84TYY514 pKa = 10.81IDD516 pKa = 4.79GLDD519 pKa = 3.54LDD521 pKa = 4.29
VV1 pKa = 7.46DD2 pKa = 4.87GEE4 pKa = 4.6PGPKK8 pKa = 9.42PGPDD12 pKa = 3.72PAPQPTPTPEE22 pKa = 3.71PTPAKK27 pKa = 9.75HH28 pKa = 6.09EE29 pKa = 4.09RR30 pKa = 11.84FIAYY34 pKa = 7.69TGTLSTLISARR45 pKa = 11.84QSSDD49 pKa = 3.46SISLYY54 pKa = 10.01SIRR57 pKa = 11.84SQRR60 pKa = 11.84IRR62 pKa = 11.84YY63 pKa = 9.1IEE65 pKa = 4.77DD66 pKa = 3.59EE67 pKa = 4.13NSSWTNIDD75 pKa = 3.62AKK77 pKa = 10.16WYY79 pKa = 8.63SQNSVEE85 pKa = 4.73AIPMFVYY92 pKa = 9.48PVPEE96 pKa = 4.3GTWSIEE102 pKa = 3.88ISCEE106 pKa = 3.95GYY108 pKa = 9.81QAASSTSDD116 pKa = 3.17PHH118 pKa = 7.88RR119 pKa = 11.84GKK121 pKa = 10.64CDD123 pKa = 2.94GMIAYY128 pKa = 9.88DD129 pKa = 4.3DD130 pKa = 4.93DD131 pKa = 4.88SSKK134 pKa = 10.55VWNVGQQNNVTITNNKK150 pKa = 9.17ADD152 pKa = 4.07NDD154 pKa = 4.0WKK156 pKa = 11.0YY157 pKa = 11.15GHH159 pKa = 7.75PDD161 pKa = 3.01LTINGDD167 pKa = 3.54RR168 pKa = 11.84FDD170 pKa = 3.77QNQVVEE176 pKa = 4.13KK177 pKa = 10.71DD178 pKa = 3.98GIISFHH184 pKa = 6.69LVTTGPNASFFLVAPAVKK202 pKa = 8.86KK203 pKa = 6.9TAKK206 pKa = 10.58YY207 pKa = 9.35NFCVSYY213 pKa = 11.32GDD215 pKa = 3.19WTDD218 pKa = 3.56RR219 pKa = 11.84DD220 pKa = 3.87MEE222 pKa = 4.49FGMVSVVLDD231 pKa = 3.47EE232 pKa = 4.66HH233 pKa = 8.3LEE235 pKa = 4.22GARR238 pKa = 11.84SSQYY242 pKa = 9.71VRR244 pKa = 11.84KK245 pKa = 7.62TPRR248 pKa = 11.84SGHH251 pKa = 5.3VGVNRR256 pKa = 11.84SHH258 pKa = 7.25RR259 pKa = 11.84LQDD262 pKa = 3.19NFVPTEE268 pKa = 3.97YY269 pKa = 11.14VSDD272 pKa = 3.89EE273 pKa = 4.39DD274 pKa = 4.06SSSNSSIVSNRR285 pKa = 11.84PSTPDD290 pKa = 3.17NDD292 pKa = 3.69SDD294 pKa = 4.09AKK296 pKa = 10.53FAEE299 pKa = 4.34SMKK302 pKa = 10.83GKK304 pKa = 10.29LPSQTKK310 pKa = 10.17LPPKK314 pKa = 10.43GFLSQLSTKK323 pKa = 8.39EE324 pKa = 3.74KK325 pKa = 11.0KK326 pKa = 10.13EE327 pKa = 3.72ISNSKK332 pKa = 8.98PSNVEE337 pKa = 3.79GLVGPLVAAYY347 pKa = 8.65GYY349 pKa = 9.02PSQTGVHH356 pKa = 6.36DD357 pKa = 4.11AARR360 pKa = 11.84EE361 pKa = 3.71ILQAKK366 pKa = 8.14EE367 pKa = 3.51AAEE370 pKa = 3.85NLAEE374 pKa = 4.64LEE376 pKa = 4.14RR377 pKa = 11.84DD378 pKa = 3.2LKK380 pKa = 10.8EE381 pKa = 4.13INKK384 pKa = 9.88LEE386 pKa = 4.37PPDD389 pKa = 4.53VIVQEE394 pKa = 4.66EE395 pKa = 4.43IPDD398 pKa = 3.87FVPPSEE404 pKa = 5.26KK405 pKa = 9.61ILKK408 pKa = 9.96EE409 pKa = 4.11DD410 pKa = 4.1DD411 pKa = 3.75PDD413 pKa = 3.72YY414 pKa = 11.5VPPIWHH420 pKa = 6.66NADD423 pKa = 3.08QAVLVSSYY431 pKa = 10.73EE432 pKa = 3.88PPDD435 pKa = 3.33WSRR438 pKa = 11.84PAYY441 pKa = 10.0EE442 pKa = 5.22SGDD445 pKa = 3.8PPKK448 pKa = 10.29KK449 pKa = 9.24TGTLKK454 pKa = 10.1GTLSKK459 pKa = 11.03LGGSLRR465 pKa = 11.84SGEE468 pKa = 4.1SSLRR472 pKa = 11.84GSLRR476 pKa = 11.84KK477 pKa = 7.81TQDD480 pKa = 2.88QTDD483 pKa = 4.12LDD485 pKa = 4.08NKK487 pKa = 10.25LSKK490 pKa = 10.77LSVIQRR496 pKa = 11.84SRR498 pKa = 11.84YY499 pKa = 8.05QRR501 pKa = 11.84ILNNLGKK508 pKa = 9.24MRR510 pKa = 11.84ARR512 pKa = 11.84TYY514 pKa = 10.81IDD516 pKa = 4.79GLDD519 pKa = 3.54LDD521 pKa = 4.29
Molecular weight: 57.7 kDa
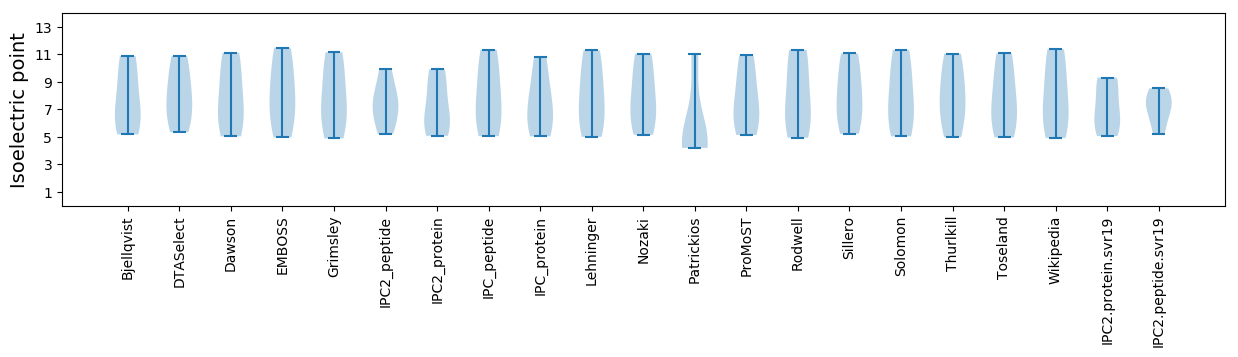
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q91QR4|Q91QR4_9LUTE RNA-directed RNA polymerase (Fragment) OS=Soybean dwarf virus OX=12049 GN=ORF2 PE=4 SV=1
MM1 pKa = 6.96VAVSNVAIQRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84TRR15 pKa = 11.84RR16 pKa = 11.84AARR19 pKa = 11.84RR20 pKa = 11.84APRR23 pKa = 11.84VQLMAVPTVTSRR35 pKa = 11.84PQRR38 pKa = 11.84RR39 pKa = 11.84GRR41 pKa = 11.84QRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84NNRR50 pKa = 11.84GGSFISGGSGKK61 pKa = 10.11AHH63 pKa = 5.8TFVFSKK69 pKa = 10.92DD70 pKa = 3.62GINGSSKK77 pKa = 10.83GSITFGPSLSEE88 pKa = 4.21CKK90 pKa = 10.0PFSDD94 pKa = 5.84GILKK98 pKa = 10.28AYY100 pKa = 10.09HH101 pKa = 6.59EE102 pKa = 4.33YY103 pKa = 10.69KK104 pKa = 9.21ITSILLQFITEE115 pKa = 4.23ASSTSSGSIAYY126 pKa = 9.43EE127 pKa = 3.79LDD129 pKa = 3.32PHH131 pKa = 6.86CKK133 pKa = 9.64YY134 pKa = 11.08SEE136 pKa = 4.03IQSLLNKK143 pKa = 10.04FSITKK148 pKa = 9.95SGSKK152 pKa = 10.08RR153 pKa = 11.84FPTRR157 pKa = 11.84AINGLEE163 pKa = 3.76WHH165 pKa = 6.97DD166 pKa = 3.68TSEE169 pKa = 4.34DD170 pKa = 3.42QFKK173 pKa = 10.28IHH175 pKa = 6.26YY176 pKa = 8.83KK177 pKa = 10.89GNGEE181 pKa = 4.19SKK183 pKa = 10.11IAGSFKK189 pKa = 10.52ISINVLTQNAKK200 pKa = 10.25
MM1 pKa = 6.96VAVSNVAIQRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84TRR15 pKa = 11.84RR16 pKa = 11.84AARR19 pKa = 11.84RR20 pKa = 11.84APRR23 pKa = 11.84VQLMAVPTVTSRR35 pKa = 11.84PQRR38 pKa = 11.84RR39 pKa = 11.84GRR41 pKa = 11.84QRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84NNRR50 pKa = 11.84GGSFISGGSGKK61 pKa = 10.11AHH63 pKa = 5.8TFVFSKK69 pKa = 10.92DD70 pKa = 3.62GINGSSKK77 pKa = 10.83GSITFGPSLSEE88 pKa = 4.21CKK90 pKa = 10.0PFSDD94 pKa = 5.84GILKK98 pKa = 10.28AYY100 pKa = 10.09HH101 pKa = 6.59EE102 pKa = 4.33YY103 pKa = 10.69KK104 pKa = 9.21ITSILLQFITEE115 pKa = 4.23ASSTSSGSIAYY126 pKa = 9.43EE127 pKa = 3.79LDD129 pKa = 3.32PHH131 pKa = 6.86CKK133 pKa = 9.64YY134 pKa = 11.08SEE136 pKa = 4.03IQSLLNKK143 pKa = 10.04FSITKK148 pKa = 9.95SGSKK152 pKa = 10.08RR153 pKa = 11.84FPTRR157 pKa = 11.84AINGLEE163 pKa = 3.76WHH165 pKa = 6.97DD166 pKa = 3.68TSEE169 pKa = 4.34DD170 pKa = 3.42QFKK173 pKa = 10.28IHH175 pKa = 6.26YY176 pKa = 8.83KK177 pKa = 10.89GNGEE181 pKa = 4.19SKK183 pKa = 10.11IAGSFKK189 pKa = 10.52ISINVLTQNAKK200 pKa = 10.25
Molecular weight: 22.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1806 |
189 |
535 |
361.2 |
40.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.423 ± 0.797 | 1.218 ± 0.327 |
5.814 ± 0.883 | 6.257 ± 0.449 |
3.931 ± 0.644 | 5.592 ± 0.774 |
2.547 ± 0.28 | 5.482 ± 0.485 |
6.921 ± 1.036 | 7.198 ± 0.492 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.547 ± 0.619 | 4.817 ± 0.104 |
5.759 ± 0.841 | 3.599 ± 0.32 |
6.478 ± 0.788 | 10.299 ± 1.488 |
4.319 ± 0.619 | 6.368 ± 0.494 |
1.107 ± 0.173 | 3.322 ± 0.127 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
